FAQ
What is the minimum volume of liquid biopsy sample that is required for profiling?
Ans: Refer to the Sample Submission Requirements Table in the ‘Service and Charges’ section.
What is the normal turnaround time for a project?
Ans: It depends mainly on the sample size of the project (no. of samples) and current availability of the team. A 100-sample project is estimated to take about 3 weeks – 1 month.
What is the acceptable quality of blood-based liquid biopsy samples for profiling?
Ans: Refer to the Haemolysis chart in the ‘Service and Charges’ section.
What are the sample types that the ncRNA core facility is capable of handling?
Ans: Refer to figure below for the list of sample types that we have worked with. For any sample types that are not listed below, we can start with a pilot feasibility study to determine the viability of the requested sample types before proceeding.
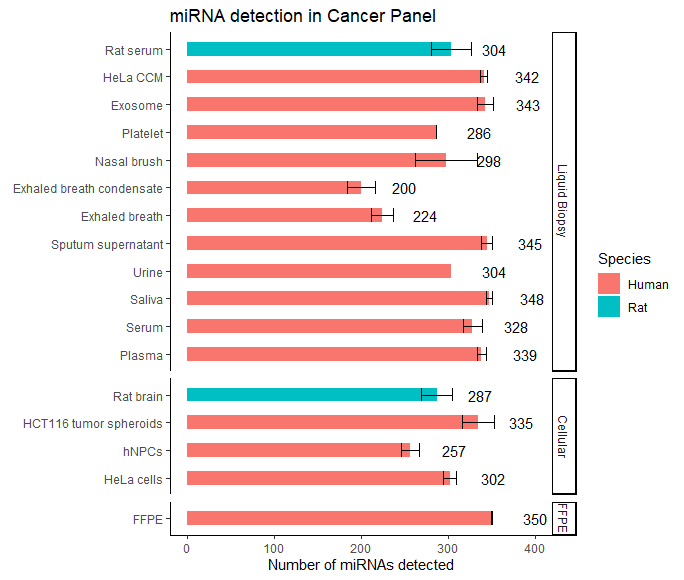
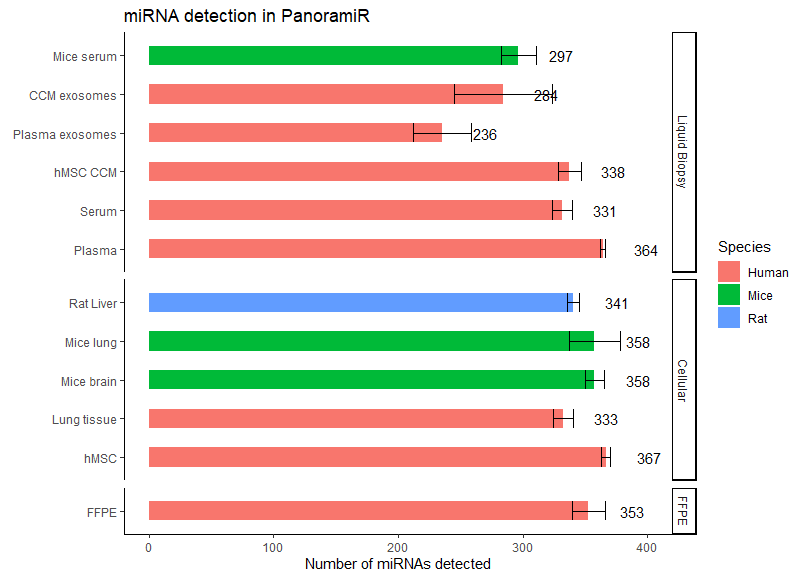
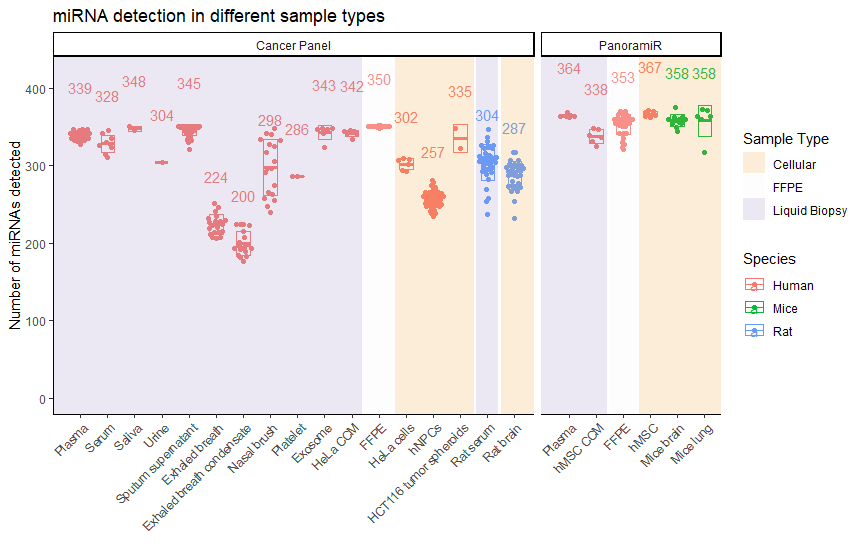
What are the types of analysis that are provided?
Ans: We will normalize the Ct values to internal controls and global mean, followed by fold change analysis. For samples with a minimum of n = 3, p-values will be calculated by Student’s t-test (for 2 group analysis) or ANOVA (for more than 3 groups analysis). We are also able to provide unsupervised hierarchical clustering heatmaps and principal component analysis (additional charges apply). Output data will include volcano plots, heatmaps and PCA plots. Please note that our standard analysis package does NOT include information on the absolute copy number (discuss with ncRNA core on how to achieve this).
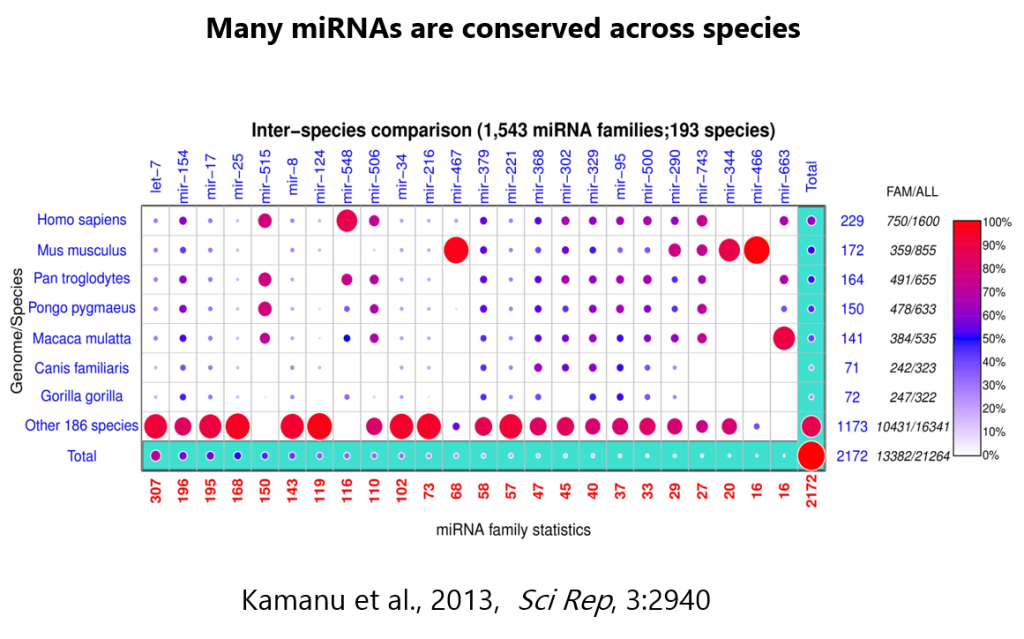
Are you able to profile the miRNAs of non-human species?
Ans: miRNA sequence conservation across species means that there is a significant overlap in the miRNAs are present in multiple species (refer to table below for a list of animal model species that we have worked with). For any species that are not listed below, we can start with a pilot feasibility study to determine the viability of the requested species before proceeding.