Noncoding RNA (ncRNA)
Reading non-coding RNAs & Decoding Life’s Secrets
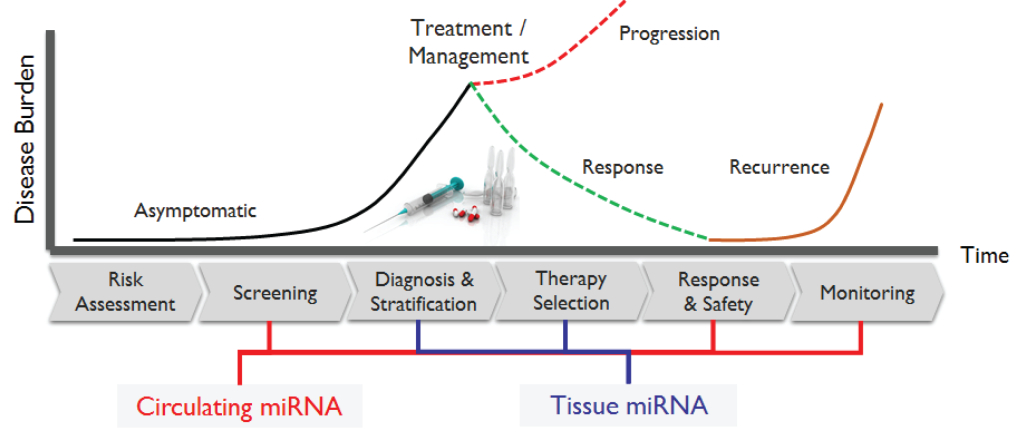
Up until recent years, RNA was implicated by Francis Crick’s central dogma as the key messenger between DNA and protein. With the advent of new technologies, we now know that the human genome encodes a vast repertoire of non-coding RNAs (ncRNAs), which was once thought to be meaningless “dark matter”. Our current understanding of ncRNAs may look like an intertwined mess of molecules, but collectively they exhibit architecture and coordination, leading to elegantly choreographed regulation of DNA and protein by RNA. While ncRNAs constitute more than 90 percent of the RNAs made from the human genome, most of the 30,000-plus known ncRNAs have been discovered in the past 10 years and are largely unstudied. To date, the most studied ncRNAs are microRNAs (miRNAs, 15-23 nucleotides long), with emerging evidence in the literature of other types of experimentally identified ncRNAs of diverse lengths and characteristics is growing steadily over the past few years.
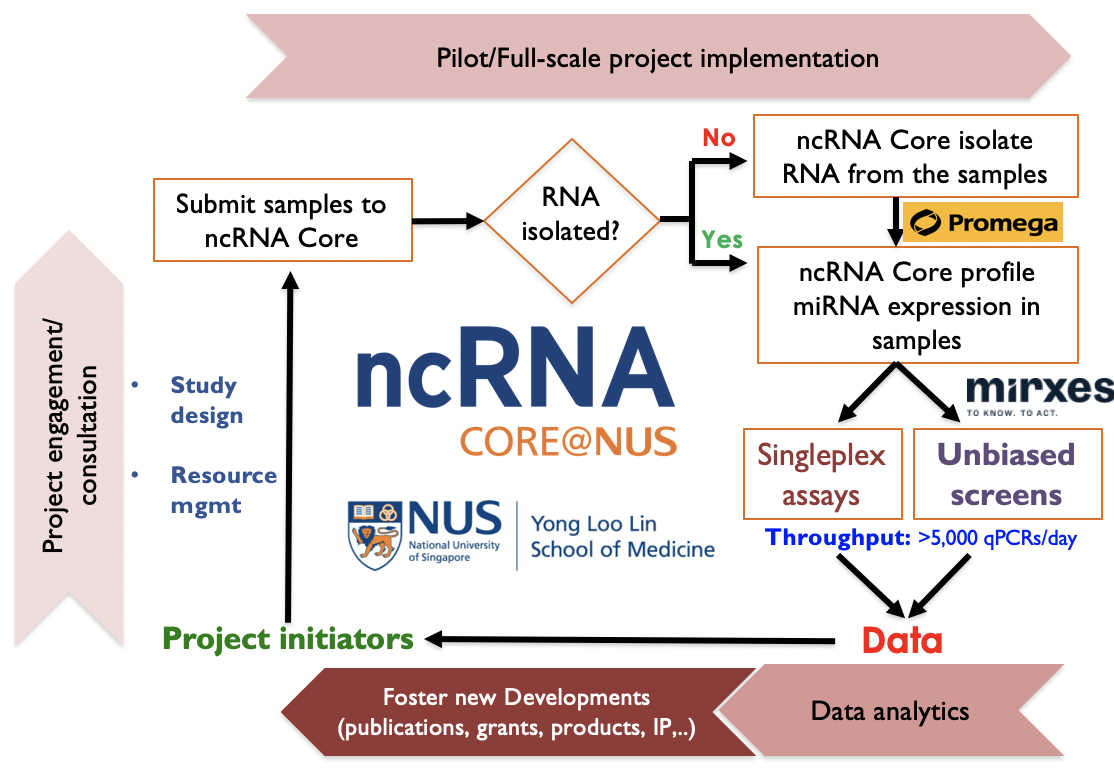
Established in 2018, the NUSMed noncoding RNA (ncRNA) Core seeks to support NUS/NUHS investigators in interrogating intracellular/circulatory ncRNA profiles/functions in diverse experimental/clinical human and animal model specimens (including biofluids, fresh frozen cells/tissues and formalin-fixed paraffin-embedded tissues). To achieve this goal, we collaborate with industry partners and leverage on their cutting edge technologies to isolate/profile/perturb ncRNAs to study their expression/forms and functions in biospecimens.
Challenges and opportunities
Check out our white paper:
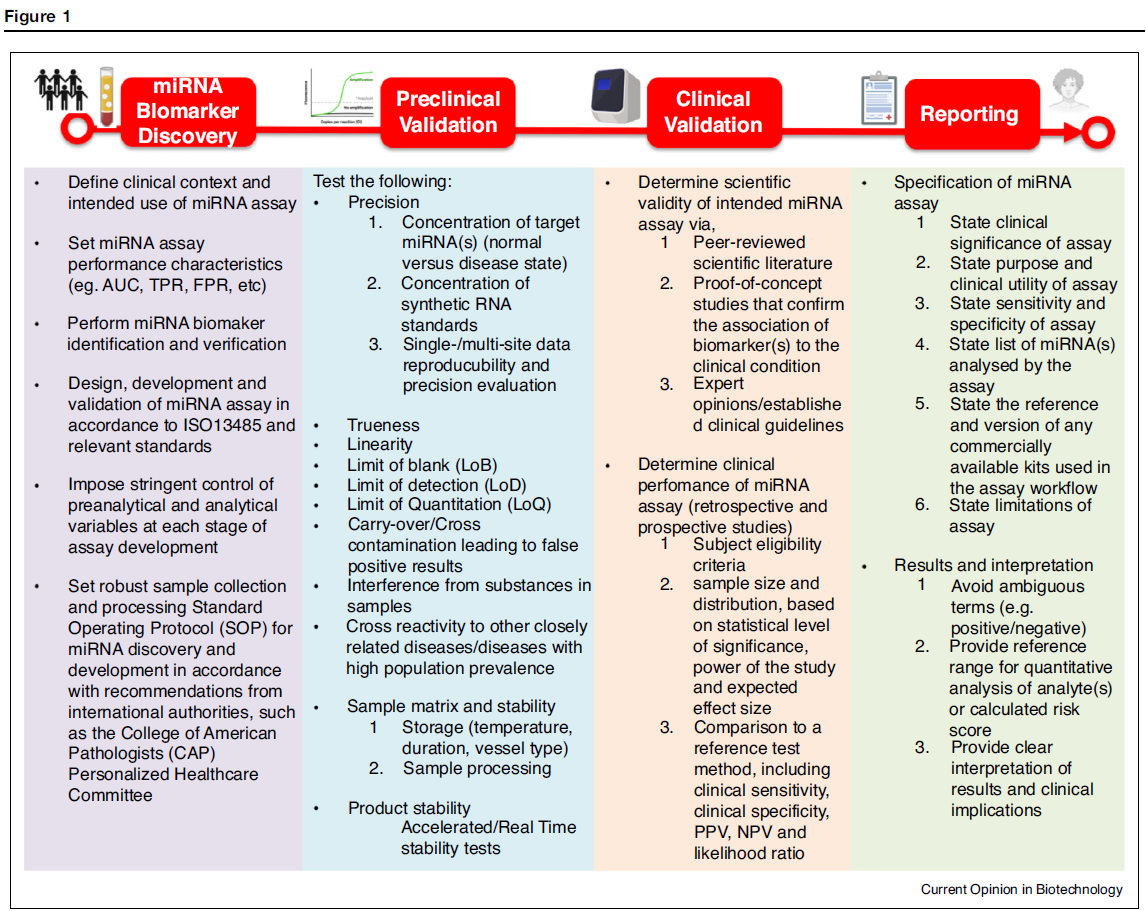
Cheong JK, Tang YC, Zhou L, Cheng H, Too HP. Advances in quantifying circulatory microRNA for early disease detection. Curr Opin Biotechnol. 2022 Apr;74:256-262. doi: 10.1016/j.copbio.2021.12.007. Epub 2022 Jan 6. PMID: 34999430.