
Issue 49
Feb 2024
A WORLD IN A GRAIN OF SAND
By Dr Volker Patzel, Department of Microbiology, Healthy Longevity Translational Research Programme, Yong Loo Lin School of Medicine, National University of Singapore
One year after the discovery of cis-splicing by the teams of Roberts and Sharp1,2, Walther Gilbert suggested that the combination of different exons of a single gene could lead to the generation of various mRNA isoforms3, a process that is today known as alternative splicing. Constitutive and alternative splicing isn’t mutually exclusive, which solves the former problem that for a continuous evolutionary process, essential genes don’t need to duplicate before one version can mutate to evolve new gene functions.
Alternative splicing generates new functions by creating new exons within existing genes and can suspend the negative selection pressure against evolutionary changes of functional genes. Thus, alternative splicing opens nearly neutral paths for evolutionary changes. Alternative splicing represents a key mechanism that is recruited by most DNA viruses, and nuclear-replicating RNA viruses to generate the full repertoire of protein functions4. RNA trans-splicing represents a special form of alternative splicing in which sequences of distinct precursor pre-mRNA transcripts are joined in trans.
In the mid-90s, first experimental evidence was obtained showing that mammalian cells can combine natural pre-mRNAs to form new mRNA via RNA trans-splicing5,6. Trans-splicing in mammalian cells differs from small leader RNA (SL RNA) trans-splicing reported earlier in trypanosomes and caenorhabditis elegans, is independent of SL RNA, and mediated by the spliceosome of the cell. Computational analyses of cDNAs from a gene databank indicated one percent of all sequenced mRNAs to be chimeric7, some of which might be synthesised by RNA trans-splicing. Up to date, several examples of naturally occurring mammalian RNA trans-splicing have been reported, many of them involving viral transcripts8-14. For many viruses, economy in genome size represents a common theme, and generating new protein functions by combining RNAs via splicing in trans does not require a genome enlargement. Our finding that the SV40 virus uses homologous RNA trans-splicing to generate a functional protein, namely a highly transforming 100 kD super T antigen, supports this hypothesis and demonstrates that viruses engage the mechanism of RNA trans-splicing to maximise their transcriptomes and proteomes15. SV40 RNA trans-splicing was found to be highly efficent with 50% or more viral transcripts being involved in the trans-splice reaction. The knowledge and understanding of the mechanism of RNA trans-splicing not only points towards a role of RNA trans-splicing in the generation of novel viral gene products, but also offers promising perspectives for various applications.
Trans-splicing based repair of gene expression
If a gene is functionally impaired by mutagenesis, gene complementation therapy may be explored to restore the defect function. Therefore, complete functional genes are delivered into target cells using viral or non-viral vectors. Many of these vectors experience cargo size limitations. If the gene complementation approach is successful, both the defect and the recombinant genes are being expressed and eventually compete for cellular functions. Sometimes, mutated genes express harmful or carcinogenic proteins.
Then, a functional gene knockdown e.g. using RNA interference is indicated which leads to the partial loss of the respective gene function. RNA trans-splicing enables repair of defect gene functions, i.e. replacement of defect with intact gene functions on the level of the pre-mRNA, thereby combing the advantages but avoiding the disadvantages of the two approaches discussed above. For applications, trans-splicing RNA (tsRNA) is designed to comprise three functional domains: a domain coding for the protein or polypeptide to be fused to the targeted gene product, a splicing domain harbouring all sequences required to recruit the spliceosome, and a binding domain (BD) (Figure a). The BD is an antisense sequence that enables the tsRNA to specifically identify the intended pre-mRNA target in the nucleus of the target cell. Once the tsRNA is bound to the pre-mRNA to edit, trans-splicing between the two RNAs occurs, triggering the generation of an edited mRNA and expression of an edited protein. Trans-splicing RNA can be designed to replace 5’ terminal, internal or 3’ terminal exons within a pre-mRNA target whereby the target RNA remains in its natural regulatory context (Figure b).
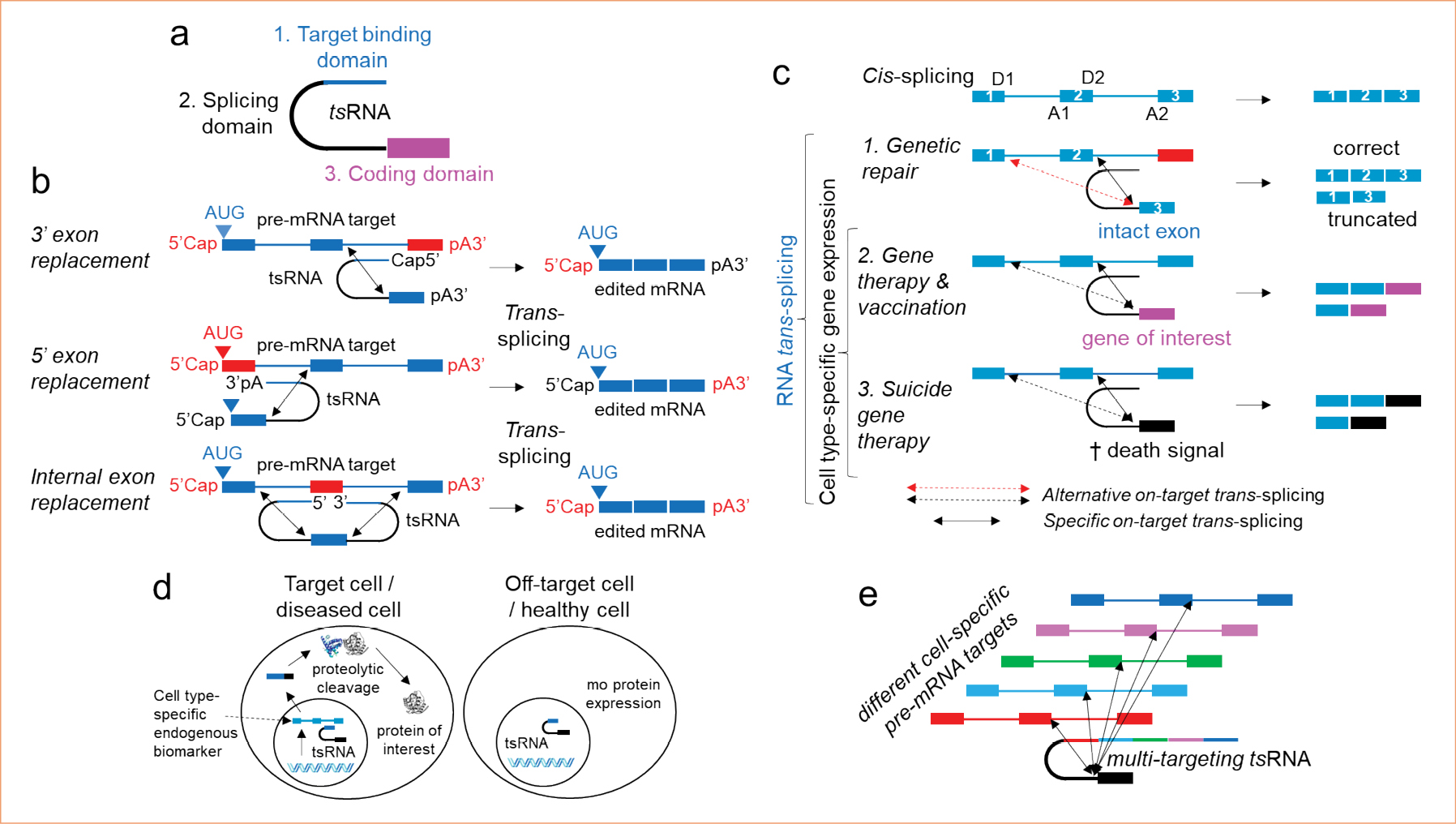
Figure. a, Design of a trans-splicing RNA (tsRNA), using the example of a tsRNA for 3‘ exon replacement. b, RNA trans-splicing mediated genetic repair via 3‘, 5‘, and internal exon replacement. c, Trans-splicing based genetic repair strategies suffer from alternative on-target trans-splicing; strategies based on cell type-specific gene expression benefit from alternative on-target trans-splicing. d, Trans-splicing triggers gene expression selectively in target/diseased cells but not in off-target/healthy cells. Initially formed chimeric proteins can efficiently be cleaved via proteolytic cleavage. e, Design of trans-splicing RNA comprising multiple target binding domains.
Opposed to RNA editing which is suitable to edit single point mutations within a pre-mRNA, trans-splicing based repair can be a more mutation agnostic approach as complete exons can be replaced. The fact that only exons but not complete genes need to be delivered to restore gene function, qualifies the trans-splicing technology to be deliverable using any vector technology, including those with apparent cargo size limitations. When compared with the CRISPR/Cas genome editing technology, RNA trans-splicing is conceptually much safer as it does not target the genomic DNA. Attempts to develop RNA trans-splicing based genetic repair therapies however have faced challenges related to achieving high on-target activity and specificity. RNA trans-splicing is competing with cis-splicing, the latter of which is favoured as it appears to be an intramolecular reaction that, unlike trans-splicing, does not require two RNAs to find each other in the nucleus of the cell. Another issue is alternative on-target trans-spicing, i.e. the fact that a trans-splicing RNA might not exclusively splice towards the intended target splice site, but in addition can also approach strong alternative splice sites leading to imprecise editing and the expression of truncated proteins (Figure c). Our studies revealed that alternative on-target trans-splicing can be 100-fold more abundant than specific on-target trans-splicing16.
Do not take roads traveled by the public.”
Cell type-specific gene expression
Because of the issues associated with trans-splicing based genetic repair, we decided to take a different path. Unlike other research teams and companies in the past, we strategised on using RNA trans-splicing to achieve cell type specificity of gene expression. Cell type specificity is a basic requirement for most gene therapy approaches as it strongly helps to reduce side effects.
Usually, it can be achieved with limited success through targeted delivery and by employing cell type-specific promoters. Intriguingly, RNA trans-splicing enables cell type-specific activation of gene expression based on the cell’s endogenous pre-mRNA biomarker profile. Therefore, the coding domain of a tsRNA can comprise the sequence coding for any gene of interest and the binding domain is designed to approach a pre-mRNA that is exclusively or predominantly expressed in the intended target cell type(s) (Figure c). Consequently, into whatever cell such a vector is being delivered, protein expression will only be triggered in target cells expressing a selected cell-specific pre-mRNA.
Resulting chimeric proteins can efficiently be cleaved via proteolytic cleavage to produce the protein of interest (Figure d). Such pre-mRNA targets have been described or can virtually be found for any target cell of interest. For many target cells, even multiple unique or overexpressed pre-mRNAs have been reported. We first designed tsRNA featured with multiple BDs each suitable to bind towards one out of a group of selected pre-mRNA targets. This design increased the trans-splicing activity 100 to 1,000 folds as it significantly increases the pre-mRNA target concentration (Figure e). In contrast to trans-splicing based genetic repair strategies, approaches to employ RNA trans-splicing for cell type-specific gene expression are not jeopardised by alternative on-target trans-splicing. On the contrary, alternative on-target trans-splicing can even amplify the level of cell type-specific expression (Figure c). For genetic repair or sustained cell specific gene expression, the tsRNA must constantly be available in the cellular nucleus which can only be achieved by a stably expressing integrating vector or the repeated delivery of a transient genetic vector.
Breaking new ground
To evade the above-mentioned delivery issues, we focused on applications that do not require sustained nuclear availability of the tsRNA. These applications include but are not limited to suicide gene therapy, whereby the expression of a death signal is specifically triggered in diseased cells, e.g. tumour or irreversibly virus-transduced cells, with the intention to kill the same without compromising healthy cells (Figure c,d)16.
Another application can be genetic vaccination. By using RNA trans-splicing, antigen expression can selectively be triggered in distinct immune cells to manipulate immune responses. We are currently investigating the suicide gene therapy and vaccination approaches in pre-clinical animal studies. In conclusion, the potential of the RNA trans-splicing technology is comparable with that of other groundbreaking RNA technologies. However, its most obvious applications and the most frequently travelled roads are not necessarily those leading to quick success. With an advanced tsRNA design and a focus on the most promising applications, RNA trans-splicing is expected to evolve into a valuable technology for gene therapeutic applications.
Berget, S. M. & Sharp, P. A. A spliced sequence at the 5′-terminus of adenovirus late mRNA. Brookhaven Symp. Biol. 29, 332–344 (1977).
Chow, L. T., Gelinas, R. E., Broker, T. R. & Roberts, R. J. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12, 1–8 (1977).
Gilbert, W. Why genes in pieces? Nature 271, 501 (1978).
Hegel R. HERNANDEZ-LOPEZ and Sheila V. GRAHAM1. Alternative splicing in human tumour viruses: a therapeutic target? Biochem. J. (2012) 445, 145–156.
Eul J, Graessmann M, Graessmann A. Experimental evidence for RNA trans-splicing in mammalian cells. EMBO J 1995; 14:3226-35.
Eul J, Graessmann M, Graessmann A. In vitro synthesized SV40 cRNA is trans-spliced after microinjection into the nuclei of mammalian cells. FEBS Lett 1996; 394:227-32.
Romani A, Guerra E, Trerotola M, Alberti S. Detection and analysis of spliced chimeric RNAs in sequence databanks. Nucleic Acids Res. 31, e17
Caudevilla C, Serra D, Miliar A, Codony S, Asins G, Bach M, et al. Natural trans-splicing in carnitin octanoyltransferase pre-mRNAs in rat liver. Proc Natl Acad Sci USA 1998; 95:12185-90.
Caudevilla C, Codony C, Serra D, Plasenica G, Roman R, Graessmann A. Localization of an exonic splicing enhancer responsible for mammalian natural trans-splicing. Nucleic Acids Res 2001; 29:3108-15.
Flouriot G, Brand H, Seraphin B, Gannon F. Natural trans-spliced mRNAs are generated from the human estrogen receptor (hER) gene. J Biol Chem 2002; 277:26244-51.
Kikumori T, Cote GJ, Gagel RF. Naturally occurring heterologous trans-splicing of adenovirus RNA with host cellular transcripts during infection. FEBS Lett 2002; 522:41-6.
Jehan Z, Vallinayagam S, Tiwari S, Pradhan S, Singh L, Suresh A. Novel noncoding RNA from human Y distal heterochromatic block (Yq12) generates testis-specific chimeric CDC2L2. Genome Res 2007; 17:423-40.
Brooks YS, Wang G, Yang Z, Smith KK, Bieberich E, Ko L. Functional pre- mRNA trans-splicing of coactivator CoAA and corepressor RBM4 during stem/progenitor cell differentiation. J Biol Chem 2009; 284:18033-46.
Desai MM, Tumurbataar S, Zhang Y, Chan LN, Sun J, Chan TS. 2011. Aberrant transcription and post-transcriptional processing of hepatitis C virus non-structural genes in transgenic mice. Transgenic Res. 20:1273-1284.
Eul J, Patzel V (2013). Homologous SV40 RNA trans-splicing: a new mechanism for diversification of viral genotypes and phenotypes. RNA Biology 10:11, 1-11, 2013.
Poddar S, Loh PS, Ooi ZH, Osman F, Eul J, Patzel V. RNA structure design improves activity and specificity of trans-splicing triggered cell death in a suicide gene therapy approach. Molecular Therapy: Nucleic Acids 11, 41-56, 2018.
More from this issue



