CRISPR’s Window to the Origins of Disease
By Dr Khor Ing Wei, Dean’s Office
Scientists have been modifying genes for decades to study their effects on biological processes. For most of this time, they had to use techniques that were laborious and not very efficient. This all changed in 2012, with the discovery that an obscure bacterial defense system, based on clustered regularly interspaced short palindromic repeats (CRISPR), could be turned into a precise, efficient and affordable tool to edit practically any genetic target.
Bacteria are thought to use CRISPR to remember and detect viruses that have infected them in the past. They do so by storing bits of genetic material from these viruses, akin to a memory bank of past enemies, in between CRISPR sequences. The bacteria also produce an RNA molecule that recognises each unique type of viral genetic material. Each RNA molecule, together with another smaller RNA, partners with an enzyme called a Cas nuclease to form a highly effective search-and-destroy team. When a virus from the memory bank is detected, the specific RNA molecule that recognises the virus’ genetic material will latch onto it, allowing Cas to cut it and stop it from becoming a new virus particle.
Scientists have fused the two RNA molecules together to form a guide RNA (gRNA) that directs Cas to a desired target.1,2 At one end of the gRNA is a spacer consisting of 20 bases (bases are the building blocks of DNA and RNA), which can be customised to bind to a desired genetic target. The other end of the gRNA binds to Cas, which cuts the target. Using this technology, genetic sequences as long as 10 kilobases can be removed and replaced with a desired sequence.3
Today, various CRISPR gene-editing kits are available for research and the technique is quickly replacing older methods such as site-directed mutagenesis, zinc finger nucleases and TALENS. Even non-scientists and schoolchildren are getting their hands wet, with a crowdfunded DIY CRISPR kit available for purchase in the U.S. (so far, it only works with bacteria).4
Here, we explore the gene editing research that is being performed at NUS Medicine, mostly using CRISPR and a form of the Cas enzyme called Cas9 (other Cas enzymes are also used around the world). This research ranges from deleting and swapping out genes to study their effects on disease processes, to developing innovative improvements to the CRISPR system.
Uncovering the mechanisms underlying diseases and treatment responses
One area where CRISPR is very useful is in generating “knockout” experimental models in which a gene or part of a gene is specifically removed and any biological changes that result from this are studied to determine the function of the gene or segment of the gene. In fact, the Transgenic and Gene Targeting Facility at the Cancer Science Institute (CSI) Singapore, headed by Dr Motomi Osato, routinely uses CRISPR-Cas9 to produce knockout models. The simple procedure involved in CRISPR gene editing enables these models to be produced in a matter of weeks, compared with the one to two years required by conventional knockout methods.
Professor H. Philip Koeffler at CSI Singapore used the CRISPR-Cas9 knockout model service at the core facility to help determine a new mechanism causing neutrophil-specific granule deficiency. In patients with this condition, the neutrophils fail to mature properly, leaving them susceptible to bacterial infections. From previous studies, the researchers knew that a protein called CCAAT/enhancer binding protein epsilon (CEBPE), was required for maturation of neutrophils and that the gene coding for CEBPE, Cebpe, was associated with neutrophil-specific granule deficiency. However, Prof Koeffler’s team did not know how the Cebpe gene was regulated. After analysing the DNA surrounding Cebpe, they found a gene segment with characteristics that made it a good candidate for a regulator of Cebpe.
Mr Yu Shuizhou and Mr Shi Jizhong at the Transgenic and Gene Targeting Facility produced a knockout model in which this gene segment, which they called the +6-kb enhancer, was selectively removed. In a paper published in Blood, Prof Koeffler and his team showed that deleting this enhancer reduced CEBPE levels and blocked the differentiation of neutrophils.5 Thus, using CRISPR gene editing, the researchers could determine that the novel enhancer played a role in blocking the differentiation of neutrophils, causing the neutrophil-specific granule deficiency condition.
Mapping the genetics and epigenetics of heart disease
Associate Professor Roger Foo, of the Department of Medicine at NUS Medicine and the National University Heart Centre Singapore, studies the molecular factors that drive the process of heart disease. Assoc Prof Foo is particularly interested in the epigenome, which refers to chemical compounds and proteins that attach to the DNA and regulate gene expression and function, such as turning a gene on or off.
Assoc Prof Foo and his team have made use of the targeting specificity of the CRISPR-Cas9 system to map out key epigenomic positions in the human heart. They are now planning to use CRISPR-Cas9 to edit the epigenome to figure out which parts are driving processes for heart disease. Once the researchers pinpoint the heart disease-relevant epigenetic changes, CRISPR-Cas9 may then be used to selectively target these areas and potentially turn specific genes on or off to treat heart disease.
Besides using CRISPR-Cas to interrogate the epigenome, Assoc Prof Foo’s team is also applying the technology to study cardiomyocytes (heart muscle cells) and the mechanisms underlying heart disease. Cardiomyocytes are difficult to extract from humans and maintain in culture. Thus, researchers have found a way to persuade pluripotent stem cells to differentiate into cardiomyocytes in the lab. However, determining the efficiency of this process can be challenging. Dr Matias Autio, a senior research fellow in Assoc Prof Foo’s lab, has designed an elegant system using CRISPR to track the differentiation of pluripotent stem cells into cardiomyocytes.
This engineered CRISPR system inserts a sequence into stem cells that consists of a fluorescent reporter gene (cerulean) as well as the MYH6 gene, which codes for the myosin heavy chain 6 (MYH6) protein. MYH6 is involved in the contraction of heart muscle; its expression in stem cells, together with other conditions, induces their differentiation into cardiomyocytes. The cerulean reporter makes the cells glow green under the microscope, indicating that the inserted gene is expressed in the cells (Figure 1). The genes are designed such that, upon expression, the cerulean reporter protein breaks off from the MYH6 protein, thus ensuring that the reporter protein does not affect the function of MYH6.
Dr Autio and his colleagues are also using CRISPR to make changes to specific nucleotide bases in genes that have been associated with heart failure. He then evaluates how cardiomyocyte function is affected by these genetic changes. This work could lead to a better understanding of the genes involved in heart diseases such as hypertrophic cardiomyopathy, in which cardiomyocytes enlarge, causing the walls of the heart to thicken and making it harder for the heart to pump blood efficiently. People with this condition may experience sudden heart attacks and death.
Making CRISPR better
Despite the power of CRISPR-Cas gene editing, the existing technology has several important limitations, including the requirement for a viral vector to deliver the guide RNA and Cas nuclease into cells. These vectors may provoke an immune response in humans, which would reduce their efficacy; they could also integrate into the host genome, causing damage or needlessly long-lasting expression. Another limitation of conventional CRISPR systems using viral vectors is that many do not work in primary cells, which are taken directly from an organism and do not multiply well when cultured in the lab.
The more Dr Volker Patzel, of the Department of Microbiology and Immunology, thought about these issues, the more he realised that the tiny DNA vectors he worked with could be better vehicles for delivering CRISPR components into cells. These dumbbell DNA vectors, so named because of their shape, are the smallest genetic expression vectors available. Their small size, and especially their tiny diameters, enable them to efficiently enter cells and the nuclei of cells. Plus, the genes that they carry can be expressed in primary cells as well as cell lines. Dumbbell vectors can be designed to induce minimal or no immune response and are safer than viral vectors because they will not integrate into the host genome.
Dr Patzel, together with his PhD student, Avantika Ghosh, have generated many different types of dumbbell vectors carrying CRISPR-Cas components (Figure 2). One of these is a 3-in-1 platform incorporating the gRNA and Cas9 gene (or, alternatively, two gRNA and the gene for the Cas9 enzyme for improved target specificity), as well as a repair template that replaces one loop of the dumbbell.
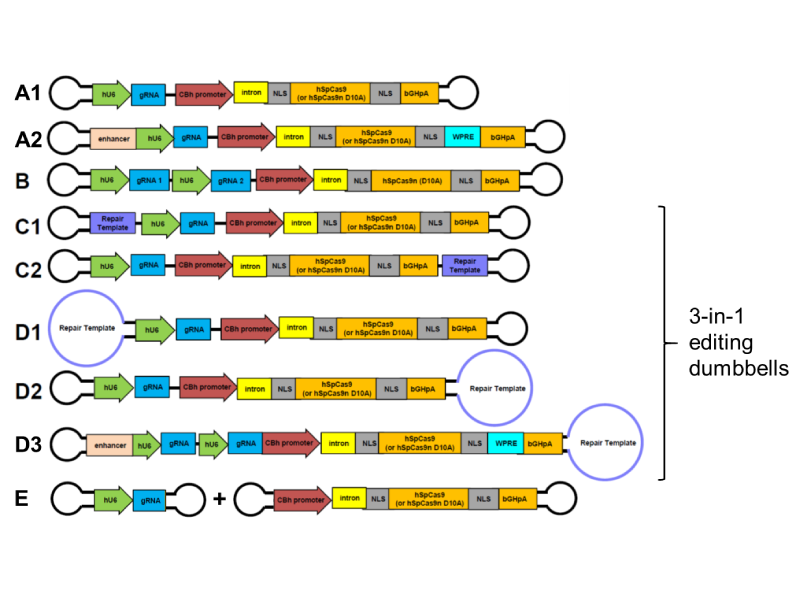
Figure 2: Different types of dumbbell vectors for introducing CRISPR-Cas components into the cell, including 3-in-1 vectors containing gRNA, Cas and repair template sequences. Image Credit: Dr Volker Patzel
When the 3-in-1 platform enters the nucleus, the gRNA and Cas9 enzyme will be expressed, forming a complex that specifically finds and cleaves the target sequence. Since the repair template is close at hand, the cell fills in the gap with a faithful reproduction of the template. Thus, the 3-in-1 CRISPR-Cas9 dumbbells are efficient, safe solutions for replacing specific genetic sequences, including repairing genetic mutations that are associated with disease. To facilitate the delivery of CRISPR-Cas9 through the nuclear membrane and to improve the gene-editing rates, some dumbbell vectors also incorporate signals that act like GPS locations, directing DNA to the nucleus (nuclear import signal) and Cas9 mRNA out of the nucleus (nuclear export signal), where it is translated to produce the Cas9 protein.
Another type of dumbbell vector that the team has developed comprises only the expression cassette for gRNA. They represent the smallest expression vectors in existence (as small as 130 base pairs), and can be finely tuned to enhance their delivery into cells. Such gRNA vectors could be used in cell lines expressing Cas9 to determine the functions of specific genes or can be combined with Cas9-expressing vectors to edit multiple targets or to optimise the amount of gRNA for efficient CRISPR gene editing.
Dr Patzel is also working on other ways to improve the usefulness and broaden the applications of the CRISPR-Cas dumbbells. For example, he and his collaborators are about to start testing the delivery of “naked” dumbbells through the skin, as well as the targeted delivery into the liver of dumbbells that have been modified with chemical moieties.
“CRISPR-Cas editing of primary cells still struggles with the lack of efficient and safe delivery vectors,” notes Dr Patzel. “Dumbbell vectors are non-integrating and not silenced in primary cells. Hence, they are most suitable to deliver the CRISPR-Cas editing technology; they do their job well and then disappear without a trace.”
CRISPR-based gene editing has come a long way in seven short years. As scientists improve the technology and address related ethical concerns, including its use in humans, and limitations, such as specificity, off-target effects and immunogenicity, it will continue to be a powerful tool for studying and treating disease.
References
1. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816-821.
2. Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Nat
Acad Sci. 2012;109:E2579-E2586.
3. Zheng Q, Cai X, Tan MH, et al. Precise gene deletion and replacement using the CRISPR/Cas9 system in human cells. Biotechniques. 2018;57:115-124.
4. Odin. DIY Bacterial Gene Engineering CRISPR Kit.
http://www.the-odin.com/diy-crispr-kit/
5. Shyamsunder P, Shanmugasundaram M, Mayakonda A, et al. Identication of a novel enhancer of CEBPE essential for granulocytic differentiation. Blood. 2019;133:2507-2517
