
Affiliations
Associate Professor (Adjunct), Department of Biochemistry, Yong Loo Lin School of Medicine, NUS
Principal Investigator, Cancer Science Institute of Singapore, NUS
Associate Director, Precision Medicine and Population Genetics (Somatic), Genome Institute of Singapore, A*STAR
Group Leader, Genome Institute of Singapore, A*STAR
Associate Professor (Adjunct), School of Biological Sciences, NTU
Biodata
Wai Leong TAM received his Bachelor’s degree from the NUS in 2003. He began his research career as a graduate student in the laboratory of Prof. Bing Lim at the Genome Institute of Singapore, where he worked on uncovering the bases for the pluripotency of embryonic stem cells and induced pluripotent stem cells. He was awarded his PhD in 2008 and continued in the same lab for a short postdoctoral stint. In 2009, Wai Leong began his postdoctoral training under the mentorship of Prof. Robert Weinberg at the Whitehead Institute in MIT, where he concentrated on understanding breast cancer stem cell biology and cancer metastasis. He joined GIS as a Principal Investigator in 2014, and his lab currently focuses on uncovering and interrogating the emerging paradigms of cancer stem cells, and designing rational strategies to specifically target cancer stem cells as a part of cancer therapy. He concurrently holds a joint appointment at the Cancer Science Institute of Singapore since 2015. In 2015, he was awarded the Singapore NRF Fellowship.
Education
| Degree and Institution | Year(s) |
| Ph.D., Stem Cell Biology, Genome Institute of Singapore | 2008 |
| B.Sc. (First Class Honors), Biology, National University of Singapore | 2003 |
Professional Experience
| Position and Institute | Year(s) |
| Principal Investigator, Cancer Science Institute of Singapore, National University of Singapore | 2015 – present |
| Adjunct Associate Professor, Department of Biochemistry, Yong Loo Lin School of Medicine, NUS | 2021 – present |
| Associate Director, Genome Institute of Singapore, A*STAR | 2021 – present |
| Group Leader, Genome Institute of Singapore, A*STAR | 2019 – present |
| Adjunct Associate Professor, School of Biological Science, NTU | 2021 – present |
| Adjunct Assistant Professor, School of Biological Science, NTU | 2017 – 2021 |
| Adjunct Assistant Professor, Department of Biochemistry, Yong Loo Lin School of Medicine, NUS | 2015 – 2021 |
| Singapore National Research Foundation Fellow, NRF | 2015 – 2020 |
| Senior Research Scientist, Genome Institute of Singapore, A*STAR | 2014 – 2019 |
| Principal Associate, Cancer Science Institute of Singapore, National University of Singapore | 2014 – 2015 |
| Postdoctoral Fellow, Laboratory of Bob Weinberg, Whitehead Institute / MIT | 2009 – 2014 |
| Research Fellow, Genome Institute of Singapore, A*STAR | 2008 – 2009 |
Research Interest
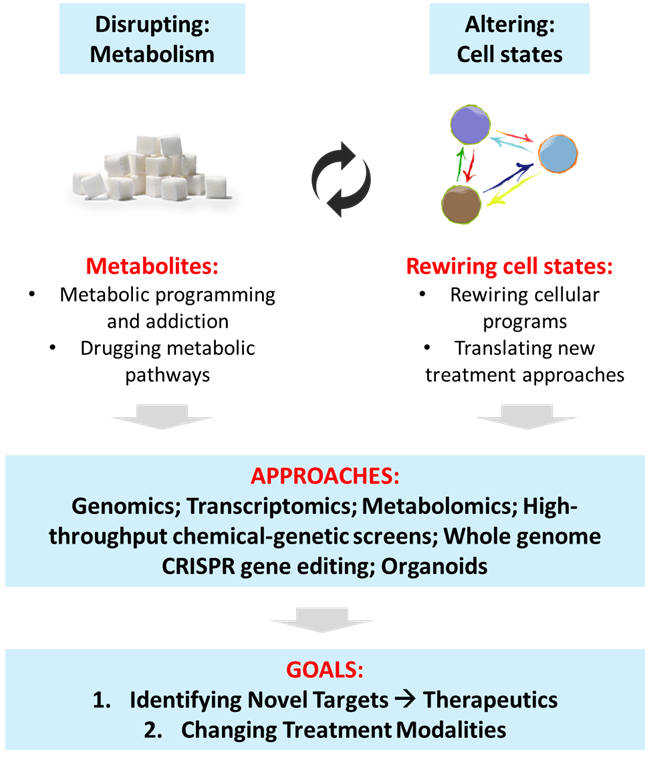
The Tam Laboratory seeks to harness functional genomics to gain insights into cancer biology for the development of selective therapeutics against cancer resistance and metastasis. We focus on dissecting two major cell-biological aspects relating to cancer stemness, resistance, and metastasis: (i) Alterations in cellular energetics; and (ii) Precise control of cell state transitions. We apply advanced methodologies that include genomics, transcriptomics, computational analyses, metabolomics, high-throughput chemical-genetic screens, genome engineering, and organoids, for building hypotheses that can lead to biological discoveries with clear translational value or intervention potential. The outcome is to achieve novel therapeutic interventions in cancer, which are effective and durable.
Research Themes
Cancer progression is orchestrated by complex alterations in gene functions and cellular behaviors. Perturbations in cell states, such as stemness vs differentiation programs, epithelial vs mesenchymal transitions, treatment resistance vs naïve responses, and localized vs metastatic phenotypes, underlie disease progression and clinical outcomes. Leveraging on genome-scale functional genomic approaches, and in close collaboration with clinician partners, we seek to gain insights into the precise control of cancer cell states, and specific targeting of metabolic pathways, for the development of selective therapeutics against cancer resistance and metastasis. We apply advanced integrative methodologies that include genomics, transcriptomics, computational analyses, metabolomics, high-throughput chemical-genetic screens, whole-genome engineering, biophysics and organoids, for enabling discoveries with clear translational or intervention value.
Cell State Transitions: Cellular heterogeneity at the genomic and transcriptomic levels is a hallmark of tumors. The heterogeneous nature of tumors may be driven by evolution of distinct clonal cell population as a result of selection or through inherent phenotypic plasticity. Cellular plasticity may be enabled by cell state transitions regulated through complex gene expression programs. Understanding cellular pathways and programs governing phenotypic plasticity will result in the more precise control of cell states and gene functions, thereby providing new opportunities to alter the course of disease. We engineer vulnerabilities into specific populations of aggressive cancer cells that will cause them to gain susceptibility to therapy, and rewire stemness and differentiation programs in cancer cells.
Cellular Energetics: Dysregulated cellular energetics occurs through the mutations in metabolism genes or loss in expression control. Several of the most effective drugs that have been in use for many decades target cancer cell metabolism. Altered metabolism is critical for tumor proliferation in general. Owing to the diverse intratumoral subpopulations that include cancer stem cells and drug resistant cells, we seek to develop more specific anti-cancer metabolic strategies for durable clinical responses, focusing on exploiting metabolic gene liabilities of cancer as therapeutic targets.
Research Themes
1. Metabolism: What are the metabolites produced and utilized by heterogeneous cancer cells? Why are they uniquely important? How do we exploit their metabolic liabilities as therapeutic targets? How do metabolic exchanges between tumor and niche cells promote disease progression?
2. Cell state plasticity: How do we engineer vulnerabilities into cancer cells that will cause them to gain susceptibility to therapy through exploiting the principles of synthetic lethality? Can we rewire stemness and differentiation programs in cancer cells? How does tumor microenvironmental cells influence cellular plasticity?
Selected Publications
For complete publication list: http://www.ncbi.nlm.nih.gov/pubmed/?term=tam+wl
- Wu Z, Tam WL. (2021) A new foe in folate metabolism. Nature Metabolism. In press. doi:10.1038/s42255-021-00474-9
- Loo SY, Toh LP, Xie WH, Pathak E, Tan W, Ma S, Lee MY, Shatishwaran S, JZZ Yeo, Yuan J, Ho YY, EKL Peh, Muniandy M, Torta F, Chan J, Tan T, Sim YR, Tan V, Tan B, Madhukumar M, Yong WS, Ong KW, Wong CY, Tan PH, Yap YS, Deng LW, Dent R, Foo R, Wenk MR, Lee SC, Ho YS, Lim EH#, Tam WL#. (2021) Fatty acid oxidation is a druggable gateway regulating cellular plasticity for driving metastasis in breast cancer. Science Advances, in press. #Corresponding authors
- Li Z, Wang Z, Lee MC, Zenkel M, Peh E, Ozaki M, Chan A, Chen S, Nakano S, Williams SEI, Orr A, Nakano M, Kobakhidze N, Zarnowski T, Popa-Cherecheanu A, Mizoguchi T, Manabe S, Hayashi K, Kazama S, Inoue K, Mori Y, Miyata K, Sugiyama K, Higashide T, Chihara E, Ideta R, Ishiko S, Yoshida A, Yanagi M, Kiuchi Y, Ohashi T, Sakurai T, Sugimoto T, Chuman H, Aihara M, Inatani M, Mori K, Ikeda Y, Ueno M, Gaston D, Bedard K, Greer WL, Chichua G, Tabagari S, Founti P, Sim KS, Chatzikyriakidou A, Pappas T, Anastasopoulos E, Lambropoulos A, Kosior-Jarecka E, Cheong A, Li A, Lukasik U, Nongpiur ME, Husain R, Wong TY, Perera SA, Álvarez L, García M, González-Iglesias H, Rodríguez-Calvo PP, Fernández-Vega L, Martinon-Torres F, Salas A, Oguz Ç, Tamcelik N, Atalay E, Batu B, Irkec M, Aktas D, Kasım B, Astakhov YS, Astakhov SY, Akopov EL, Emelyanov A, Vysochinskaya V, Uebe S, Krumbiegel M, Welge-Luessen U-C, Mardin C, Berner D, Hoja U, Kruse FE, , Reis A, Moebus S, Carmichael TR, Hauser M, Ramsay M, Mossböck G, Nilgun Y, Tashiro K, Konstas AGP, Coca-Prados M, Foo JN, Sotozono C, Kubota T, Dubina M, Topouzis F, Ritch R, Pasutto F, Wiggs J, Schloetzer-Schrehardt U#, Ho YS#, Aung T#, Tam WL#, Khor CC#. Association of rare CYP39A1 variants with exfoliation syndrome involving the anterior chamber of the eye. (2021) Journal of the American Medical Association. 22 Feb. #Corresponding authors
- Sun J, Prabhu N, Tang J, Yang F, Jia L, Guo J, Xiao K, Tam WL#, Nordlund P#, Dai L#. (2021) Recent advances in proteome-wide label-free target deconvolution for bioactive small molecules. Medicinal Research Reviews. 2021 Feb 3. doi: 10.1002/med.21788. #Corresponding authors
- Nguyen PHD, Ma S, Phua CZJ, Kaya NA, Lai HLH, Lim CJ, Lim JQ, Wasser M, Lai L, Tam WL, Lim TKH, Wan WK, Loh T, Leow WQ, Pang YH, Chan CY, Lee SY, Cheow PC, Toh HC, Ginhoux F, Iyer S, Kow AWC, Young Dan Y, Chung A, Goh BKP, Albani S, Chow PKH, Zhai W, Chew V. Intratumoural immune heterogeneity as a hallmark of tumour evolution and progression in hepatocellular carcinoma. (2021) Nature Communications. 12(1):227. doi: 10.1038/s41467-020-20171-7.
- Chen, J., Zhang, T., Yang, H., Teo, A.S.M., Tan, C.Q., Lu, B., Alvarez, J.J.S., Lim, J.Q., Chan, C.X., Faranak, G.S., Takano, A., Nahar, R., Lee, Y.Y., Chua, K.P., Lim, C.H., Koh, T.P.T, Aung, Z.W., Lim, T.K.H, Wilm, A., Liew, A.A., Lau, D.P.X., Kwang, X.L., Toh, C.K., Lim, W.T., Lim, B., Liu, J., Tam, W.L., Tan, E.H., Creasy, C., Tan, D.S.W., Hillmer, A.M., and Zhai, W. (2020) The genomic landscape and ethnic specificity of lung Adenocarcinoma in Asia. Nature Genetics, 52(2):177-186.
- Tan, J.L., Li, F., Yeo, J.Z., Yong, K.J., Bassal, M.A., Ng, G.H., Lee, M.Y., Leong, C.Y., Tan, H.K., Wu, C.S., Liu, B.H., Chan, T.H., Tan, Z.H., Chan, Y.S., Wang, S., Lim, Z.H., Toh, T.B., Hooi, L., Low, K.N., Ma, S., Kong, N.R., Stein, A.J., Wu, Y., Thangavelu, M.T,, Suzuki, A., Periyasamy, G., Asara, J.M., Dan, Y.Y., Bonney, G.K., Chow, E.K., Lu, G.D., Ng, H.H., Kanagasundaram, Y., Ng, S.B., Tam, W.L.#, Tenen, D.G.#, and Chai, L.# (2019) New High-throughput Screen Identifies Compounds That Reduce Viability Specifically In Liver Cancer Cells That Express High Levels of SALL4 by Inhibiting Oxidative Phosphorylation. Gastroenterology. Aug 22. pii: S0016-5085(19)41242-0. doi: 10.1053/j.gastro.2019.08.022. #Corresponding authors
- Kim, J.J., Lee, Y.A., Su, D., Lee, J., Park, S.J., Kim, B., Lee, J.H.J., Lee, J.S., Hong, S.C., Wang, L., Samanta, A., Kwon, H.Y., Kim, J.Y., Yu, Y.H., Ha, H.H., Wang, Z., Tam, W.L., Lim, B., Kang, N.Y., Chang, Y.T. (2019) A NIR probe tracks and treats lung tumor initiating cells by targeting HMOX2. Journal of the American Chemical Society. Aug 22. doi: 10.1021/jacs.9b06068
- Wang, Z., Yip, L.Y., Lee, J.H.J., Wu, Z., Chew, H.Y., Chong, P.K.W., Teo, C.C., Ang, H.Y.K, Peh, K.L.E., Yuan, J., Choo, L.S.K., Basri, N., Jiang, X., Yu, Q., Hillmer, A., Lim, T.K.H, Takano, A., Tan, E.H., Tan, D.S.W., Ho, Y.S., Lim, B., and Tam, W.L. (2019) Methionine is a metabolic dependency of tumor-initiating cells. Nature Medicine. 25(5):825-837.
- Lai, G.G.Y., Lim, T.H., Lim, J., Liew, P.J.R., Kwang, X.L., Nahar, R., Aung, Z.W., Takano, A., Lee, Y.Y., Lau, D.P.X., Tan, G.S., Tan, S.H., Tan, W.L., Ang, M.K., Toh, C.K., Tan, B.S., Devanand, A., Too, C.W., Gogna, A., Ong, B.H., Koh, T.P.T., Kanesvaran, R., Ng, Q.S., Jain, A., Rajasekaran, T., Yuan, J., Lim, T.K.H., Lim, A.S.T., Hillmer, A.M., Lim, W.T., Iyer, N.G., Tam, W.L., Zhai, W., Tan, E.H., and Tan, D.S.W. (2019) Clonal MET Amplification as a Determinant of Tyrosine Kinase Inhibitor Resistance in Epidermal Growth Factor Receptor-Mutant Non-Small-Cell Lung Cancer. Journal of Clinical Oncology. 37(11):876-884.
- Lee, Y.-A., Kim, J.-J., Lee, J., Sahu, S., Kwon, H.-Y., Park, S.-J., Lee, J.H.J, Jang, S.-Y., Lee, J.-S., Wang, Z., Tam, W.L., Lim, B., Kang, N.-Y., Chang, Y.T. (2018) Identification of Tumour initiating cells by a small molecule fluorescent probe through vimentin as the biomarker. Angewandte Chemie International Edition. 57(11):2851-2854.
- Nahar, R., Zhai, W., Zhang, T., Takano, A., Khng, A.J., Lee, Y.Y., Liu, X., Lim, C.H., Koh, T.P.T., Aung, Z.W., Lim, T.K.H., Veeravalli, L., Yuan, J., Teo, A.S.M., Chan, C.X., Poh, H.M., Chua, I.M.L., Liew, A.A., Lau, D.P.X., Kwang, X.L., Toh, C.K., Lim, W.-T., Lim, B., Tam, W.L., Tan, E.H., Hillmer, A.M., Tan, D.S.W. (2018) Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nature Communications. 9(1):216.
- Goh, J.Y., Feng, M., Wang, W., Oguz, G., Yatim, S.M.J.M., Lee, P.L., Bao, Y., Lim, T.H., Wang, P., Tam, W.L., Kodahl, A.R., Lyng, M.B., Sarma, S., Lin, S.Y., Lezhava, A., Yap, Y.S., Lim, A.S.T., Hoon, D.S.B., Ditzel, H.J., Lee, S.C., Tan, E.Y., Yu, Q. (2017) Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nature Medicine. 23(11):1319-1330.
- Tan, W.L., Jain, A., Takano, A., Newell, E.W., Iyer, N.G., Lim, W.T., Tan, E.H., Zhai, W., Hillmer, A.M., Tam, W.L., and Tan, D.S.W. (2016) Novel therapeutic targets on the horizon for lung cancer. Lancet Oncology. 17:e347
- Zhang, W.C., Chin, T.M., Yang, H., Nga, M.N., Lunny, D.P., Lim, E.K.H., Sun, L.L., Pang, Y.H., Leow, Y.N., Malusay, S.R.Y., Lim, P.X.H., Lee, J.Z., Tan, B.J.W., Shyh-Chang, N, Lim, E.H., Lim, W.T., Tan, D.S.W., Tan, E.H., Tai, B.C., Soo, R.A., Tam, W.L.# and Lim, B.# (2016) Tumor-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nature Communications. 7:11702. #Corresponding authors
- Pattabiraman, D.R., Bierie, B., Kober, K., Krall, J., Zill, C., Reinhardt, F., Tam, W.L. and Weinberg, R.A. (2016). cAMP-induced activation of PKA leads to epigenetic reprogramming-mediated mesenchymal-to-epithelial transition and exit from the cancer stem cell state. Science. 351:6277
- Ye, X., Tam, W.L., Shibue, T., Kaygusuz, Y., Reinhardt, F., Eaton, E. Weinberg, R.A. (2015). Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 525:256-60.
- Lu, H., Clauser, K.R., Tam, W.L., Frose, J., Reinhardt, F., Baty, C.J., Donnenberg, V.S., Carr, S.A., Weinberg, R.A. (2014). A cancer-stem-cell niche formed by juxtacrine signaling between tumor-associated macrophages and breast cancer stem cells. Nature Cell Biology. 16:1105-17.
- Shaul, Y.D., Freinkman, E., Comb, W.C., Cantor, J.R., Tam, W.L., Thiru, P., Kim, D., Pacold, M.E., Chen, W.W., Bierie, B., Possemato, R., Weinberg, R.A., Yaffe, M.B. and Sabatini, D.M. (2014). DPYD is a key component of a metabolic gene expression program required for the epithelial-mesenchymal transition. Cell. 158:1-16.
- Tam, W.L. and Ng, H.H. (2014). Sox2: Masterminding the root of cancer. Cancer Cell. 26:3-5.
- Chen, X., Iliopoulos, D., Zhang, Q., Tang, Q., Greenblatt, M.B., Hatziapostolou, M., Lim, E., Tam, W.L., Ni, M., Chen, Y., Mai, J., Shen, H., Hu, D.Z., Adoro, S., Hu, B., Song, M., Landis, M.D., Ferrar, m., Brown, M., Chang, J.C., Liu, X.S., and Glimcher, L.H. (2014). XBP1 promotes human triple negative breast cancer by controlling the hypoxia response. Nature. 508:103-107.
- Tam, W.L. and Weinberg, R.A. (2013). The epigenetics of epithelial-mesenchymal plasticity in cancer. Nature Medicine. 19:1438-1449
- Tam, W.L., Lu, H., Buikhuisen, J., Soh, B.S., Lim, E., Reinhardt, F., Wu, Z.J., Krall, J.A., Bierie, B., Guo, W., Chen, X., Liu, X.S., Brown, M., Lim, B. and Weinberg, R.A. (2013). Protein kinase C α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell. 24:347-364
- Guo, W., Keckesova, Z., Donaher, J.L., Shibue, T., Reinhardt, F., Itzkovitz, S., Bell, G., Tam, W.L., Tischler, V., Mani, S.A., van Oudenaarden, A., and Weinberg, R.A. (2012). Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 148:1015-1028
- Han, J., Yuan, P., Yang, H., Zhang, J., Soh, B.S., Li, P., Lim, S.L., Cao, S.Y., Tay, J.L., Orlov, Y.L., Lufkin, T., Ng, H.H., Tam, W.L.# and Lim, B.# (2010). Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 463:1096-1100. #Corresponding authors
- Lim, C.Y.*, Tam, W.L.*,#, Zhang, J.*, Ang, H.S, Jia, H., Lipovich, L., Ng, H.H., Wei, C.L., Sung, W.K., Robson, P., Yang, H. and Lim, B.# (2008). Sall4 regulates distinct transcription circuitries in different embryo-derived stem cell lineages. Cell Stem Cell. 3:543-554. *Equal contributions; #Corresponding authors
- Tam, W.L., Lim, C.Y., Han, J., Zhang, J., Ang, Y.S., Ng, H.H., Yang, H.H. and Lim, B. (2008). Tcf3 regulates embryonic stem cell pluripotency and self-renewal by the repression of Oct4 and multiple lineage pathways. Stem Cells. 26:2019-2031
- Tay, Y.M.S.*, Tam, W.L.*, Ang, Y.S.*, Gaughwin, P.M., Wang, W.J., Liu, R., George, J., Ng, H.H., Miranda, K.C., Perera, R.J., Lufkin, T., Rigoutsos, I., Thomson, A. and Lim, B. (2008). MicroRNA-134 modulates the differentiation of mouse embryonic stem cells. Stem Cells. 26: 17-29. *Equal contributions
- Zhang, J.*, Tam, W.L.*, Tong, G.*, Wu, Q., Chan, H.Y., Lufkin, T., Soh, B.S., Lou, Y., Ng, H.H., Robson, P and Lim, B. (2006). Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Oct4. Nature Cell Biology. 8: 1114-1123. *Equal contributions
- Miranda K.C., Huynh T., Ang, Y.S., Tay Y., Tam, W.L., Thomson A.M., Lim B. and Rigoutsos I. (2006). A pattern-based method for the identification of microRNA-target sites and their corresponding RNA/RNA complexes. Cell. 126: 1203-1217

