
Affiliations
Associate Professor, Department of Biochemistry, Yong Loo Lin School of Medicine, NUS.
Co-Director, Infectious Diseases Translational Research Program, Yong Loo Lin School of Medicine, NUS.
Assistant Dean, Equal Opportunities and Career Development, Yong Loo Lin School of Medicine, NUS.
Biodata
Yunn-Hwen Gan graduated from Purdue University with a B.Sc. (Honours) in Molecular Biology and University of Wisconsin-Madison with a Ph.D. in medical microbiology and immunology. She is a world leading researcher in melioidosis, a disease primarily in the tropics caused by the bacterium Burkholderia pseudomallei. She established the first mucosal animal model in 2002 and had helped define the field by examining pathogen’s virulence as well as the host immune response to the disease. Her lab discovered a regulatory cascade that coordinately controlled two bacterial secretion systems which are absolutely critical for bacterial virulence. Other seminal findings include the discovery that glutathione deficiency is the reason for increased disease risk to melioidosis for Type 2 diabetic patients.
Her current research focuses on Klebsiella induced liver abscess, a prominent disease in Asia, particularly in regions of China, Taiwan, Hong Kong, Singapore and South Korea. Her work involves identifying bacterial virulence factors of hypervirulent Klebsiella pneumoniae responsible for causing liver abscess. Her team examines host and bacterial factors that affect gut colonization and translocation. Her recent works have identified the alarming trend of the convergence of multidrug resistance and hypervirulence in K. pneumoniae in Singapore’s hospital settings. Her ongoing research investigates how antibiotic resistance genes on highly evolved and adapted plasmids dominant in clinical bacterial isolates are spreading among bacterial populations, and strategies to stop the spread.
She has also established multidisciplinary collaborations with chemists, clinicians and computational biologists to examine novel strategies to treat antibiotic resistant Enterobacteriaceae bacteria. One of such strategies is to establish a synthetic commensal community of bacteria to be used as probiotics for decolonization from the gut. Another strategy is to partner with AI scientists to employ synergy testing of FDA-approved compounds, including those with no known antibacterial activity on their own, on multidrug resistant bacteria.
Research Interest
- Novel strategies for targeting multidrug resistant enteric bacteria.
- Multidrug resistance plasmid transmission in clinical Gram-negative bacteria.
- Bacterial sensing of host environment by two component sensor/regulator systems.
- Identifying risk factors for Klebsiella induced liver abscess.
- The influence of microbiome on Klebsiella pneumoniae gut colonization
Projects
Klebsiella Induced Liver Abscess (KLA)
KLA is the single most important cause of monomicrobial liver abscess and its subsequent complications in many parts of Asia, including Singapore. We examine bacterial factors and the ensuing host immune responses in the encounter between hypervirulent and its mammalian host. It is believed that many infections initiate from bacteria colonized in the intestines as they translocate across the intestinal barrier and travel via the hepatic portal vein to the liver. We are interested in examining both bacterial and host factors at play at each stage of the infection, including during the colonization stage. We routinely use a hypervirulent strain isolated from a liver abscess patient designated as a reference strain, SGH10, for our investigations. Using murine colonization and infection models, as well as primary human intestinal organoids, we hope to understand when and how the bacteria transition from a gut colonizer to a potentially deadly pathogen.
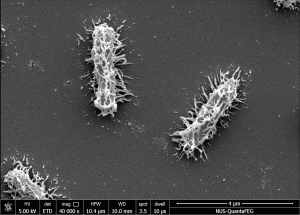
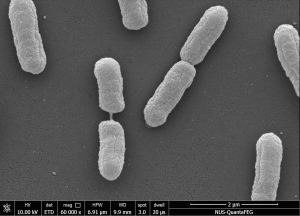
Figure 1. Scanning Electron Microscope (SEM) images of SGH10 WT strain at 40,000X magnification (left) and SGH10ΔwcaJ capsule mutant strain at 60,000X magnification (right). (Publication link : 🔗)
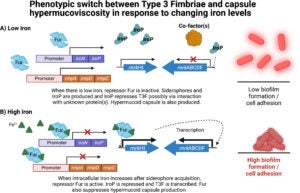
Figure 2. Transcriptional phenotypic switch between T3F and capsule hypermucoviscosity in response to changing iron levels. (A) Low iron condition results in hypermucoid capsule production and repressed T3F, leading to low biofilm formation and cell adhesion. (B) Iron-rich condition results in expression of T3F and downregulation of capule mucoviscosity, leading to high biofilm formation and cell adhesion. (Publication link : 🔗)

Figure 3. Genomic Islands GIE492 and ICEKp10 enable hypervirulent K. pneumoniae to kill several commensal bacterial taxa during interspecies interactions in the gut. Thus, acquisition of GIE492 and ICEKp10 could enable better carriage in host populations and explain the dominance of the CG23-I HvKp lineage. (Publication link : 🔗)
Multidrug resistance plasmid transmission in Klebsiella pneumoniae
We are interested in understanding the factors driving the transmission of carbapenem-resistance and multi-drug resistance plasmids in hospitals. We track plasmid and resistance genes dominance over time in a multi-centre collaboration, then apply molecular genetics to determine plasmid and bacterial host factors favouring spread and stability of carriage. In particular, we focus on the emergence of hypervirulent and carbapenem-resistant K. pneumoniae strains and aim to define factors and conditions leading to this convergence.
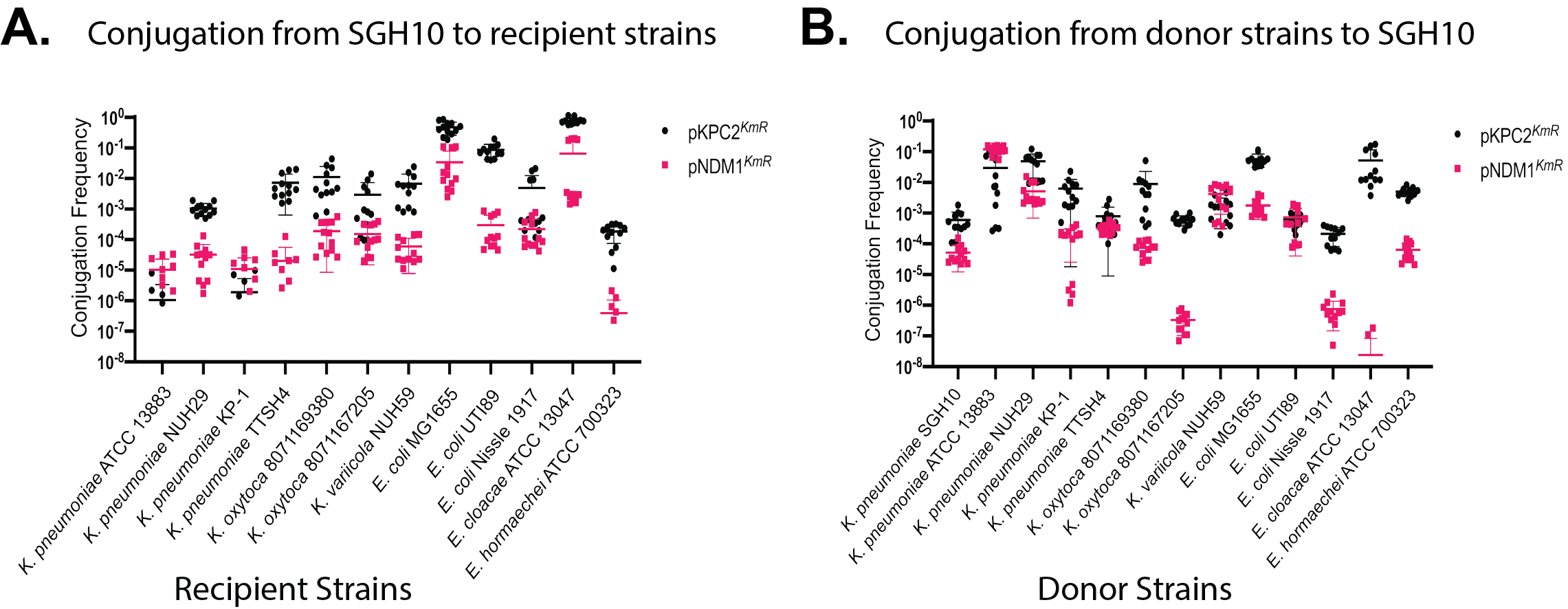
Figure 4. Conjugation frequency of pKPC2 and pNDM1 among various Enterobacterales donor-recipient pairs. (A) Conjugation frequency of pKPC2 and pNDM1 from K. pneumoniae SGH10 plasmid donor strain to a panel of Enterobacterales recipient strains; and B) from the panel of Enterobacterales plasmid donor strains to K. pneumoniae SGH10 recipient strain. Each symbol represents 1 experimental replicate with a total of 12 replicates. Data points that are not seen on the graphs indicate no detectable transconjugant. (Publication link : 🔗)
Novel strategies to target transmission of carbapenem-resistance in Enterobacterales
We engage in extensive collaborations with clinical colleagues, computational biologists, and chemists to develop strategies for combating carbapenem resistant Enterobacterales (CRE). Our efforts involve evaluating the efficacy and clinical potential of innovative synthetic antibacterial compounds and materials. Furthermore, we explore combinatorial therapy guided by AI platform to identify novel drug combinations capable of effectively eradicating CRE.

Figure 5. OIM1-6’s DNA binding properties and resulting DNA damage. (A) Flow cytometry analysis demonstrates picogreen displacement by OIM1-6 inside bacterial cytoplasm, indicating the DNA-binding properties of OIM1-6. Picogreen is a DNA binding dye with fluorescence level measured under Alexa Fluor 488 channel. (B) Confocal images showing the treatment of OIM1-6 leads to more foci formation in E. coli Gam-GFP strain as compared to the untreated cells. The hydrogen peroxide cells served as positive control with elevated level of foci formation. The presence of foci is indicative of double-stranded DNA breaks caused by the drug treatment. (Publication link : 🔗)
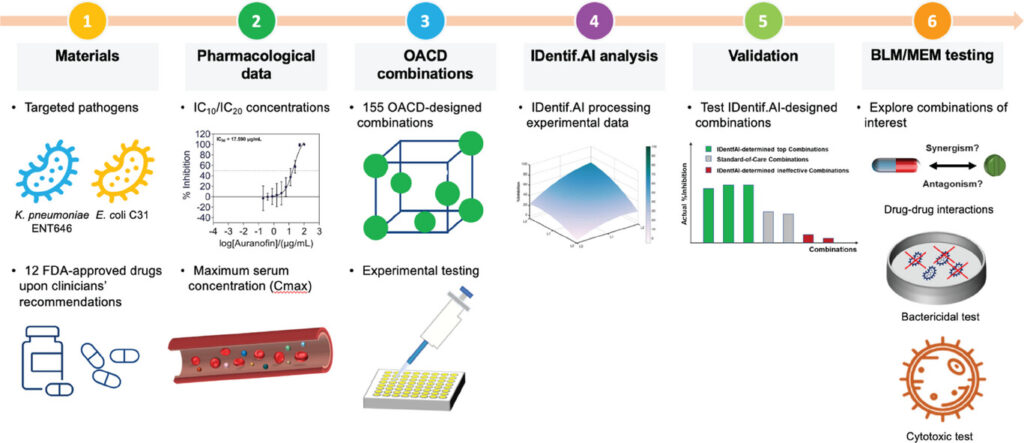
Figure 6. Workflow of the IDentif.AI-guided drug combinatorial study against CRE. Two clinical isolates from the most representative CRE species (hvKp ENT646 and E. coli C31) are investigated by IDentif.AI. The workflow begins by selecting 12 FDA-approved drugs, and they are individually assessed via dose-response experiment in vitro. Relevant concentration levels are selected. Subsequently, 155 OACD-designed combinations are experimentally validated, and the respective data are analyzed by IDentif.AI. Top combinations selected by the platform are comprehensively analyzed. (Publication link : 🔗)
Interaction of Burkholderia pseudomallei with host cells
B. pseudomallei is unique among bacterial pathogens in its ability to fuse host cells into multinucleated giant cells (MNGCs). Fusion is dependent on bacterial intracellular motility through the polymerization of actin into “actin comet tails” and the Type 6 Secretion System (T6SS5). We discovered that the process of cell fusion results in genomic instability and formation of micronuclei. Host cells sense danger brought on by unnatural cell fusion and this then triggers the cGAS-STING innate immune signalling pathway that ultimately leads to autophagic cell death. This could be the body’s defense against cellular transformation.
One of the routes of infection is through breaks in skin, such as those experienced by farmers with exposure to soil. We also examine the interaction of the bacteria with primary keratinocytes and the response of the host at this encounter.
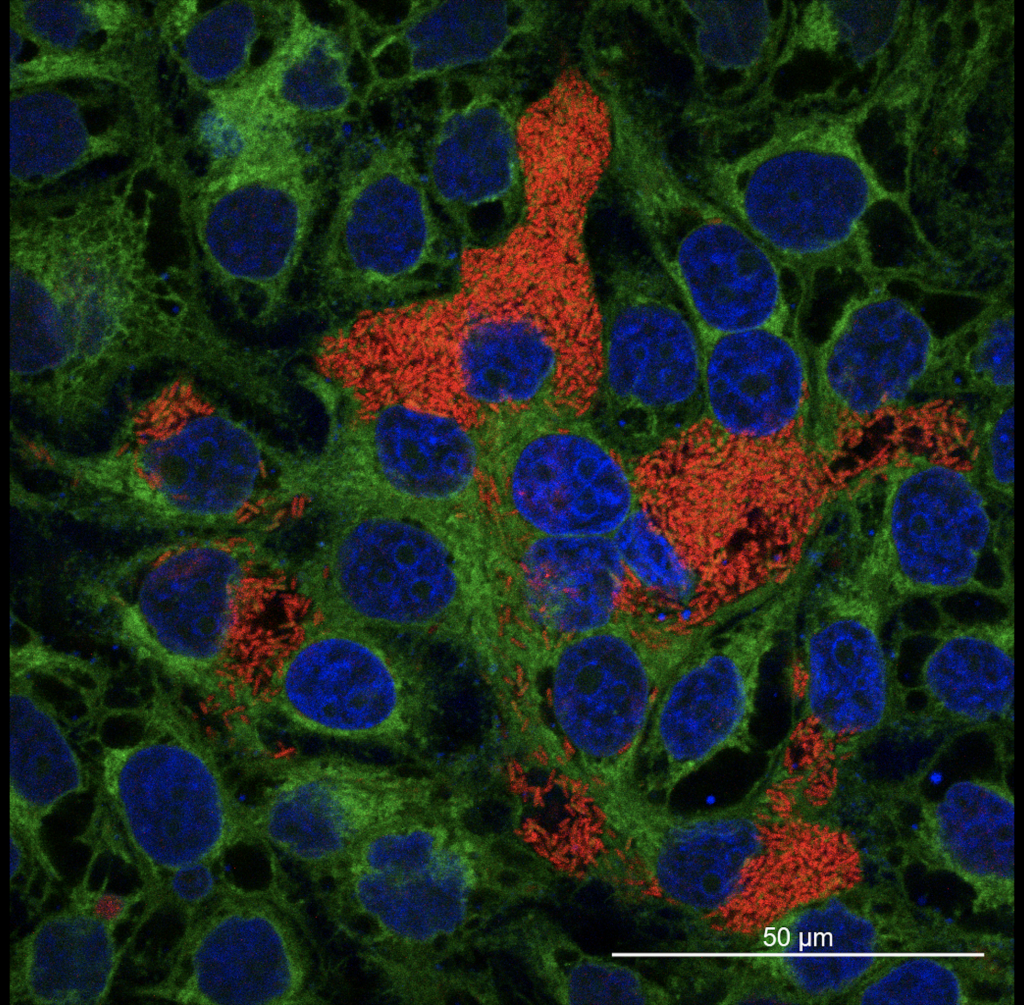
Figure 7. MNGC formation in HepG2 liver epithelial cell. Bacteria (Burkholderia spp.) is shown in red, the cell periphery in green and the cell nuclei in blue. (Publication link : 🔗)
Movie 1. B. thailandensis infected multinucleated giant cell containing highly condensed mitotic-like chromosomes. Cell rounds up, initiates cytokinesis but eventually abort the process. HepG2 cells were infected with mApple fluorescent B. thailandensis (in Red) and stained with CellMask plasma membrane dye (in Magenta) and Hoechst 33342 DNA dye (in Blue).
Publications
-
Oo G, Low WW, Yong M, Stanton TD, Ayuni NN, Bifani P, Wyres KL, Gan YH. Anti-plasmid defense in hypervirulent Klebsiella pneumoniae involves Type I-like and Type IV restriction modification systems. Emerg Microbes Infect. 2025 Dec;14(1):2558877. doi: 10.1080/22221751.2025.2558877. Epub 2025 Sep 22. PMID: 40916842; PMCID: PMC12456054. 🔗
-
Koh CH, Lambu MR, Tan C, Wei G, Kok ZY, Zhang K, Vu QHN, Panneerselvam M, Ooi YJ, Tan SH, Wang Z, Tatina MB, Ng JTY, Guo A, Tonanon P, Dang TT, Gan YH, Mu Y, Hammond PT, Chi YR, Webster RD, Pullarkat SA, Li Q, Greenberg EP, Gründling A, Pethe K, Chan-Park MB. Carbene formation as a mechanism for efficient intracellular uptake of cationic antimicrobial carbon acid polymers. Nat Commun. 2025 Jul 12;16(1):6460. doi: 10.1038/s41467-025-61724-y. PMID: 40652000; PMCID: PMC12255753. 🔗
-
Lim C, Zhang CY, Cheam G, Chu WHW, Chen Y, Yong M, Lim KYE, Lam MMC, Teo TH, Gan YH. Essentiality of the virulence plasmid-encoded factors in disease pathogenesis of the major lineage of hypervirulent Klebsiella pneumoniae varies in different infection niches. EBioMedicine. 2025 May;115:105683. doi: 10.1016/j.ebiom.2025.105683. Epub 2025 Apr 4. PMID: 40184910; PMCID: PMC12002934. 🔗
-
Si Z, Sun Y, Tan C, Ooi YJ, Li M, Raju C, Shubi J, Gan YH, Zhu Y, Li P, Chan-Park MB, Pethe K. A cationic main-chain poly(carbonate-imidazolium) potent against Mycobacterium abscessus and other resistant bacteria in mice. Biomaterials. 2025 May;316:123003. doi: 10.1016/j.biomaterials.2024.123003. Epub 2024 Dec 12. PMID: 39709850. 🔗
-
Shorey S, Gan YH, Cavert MS, Archuleta S. Is medical school culture conducive to women’s academic success? a survey on faculty perceptions and experiences of gender equity. BMC Med Educ. 2024 Dec 18;24(1):1462. doi: 10.1186/s12909-024-06470-3. PMID: 39696357; PMCID: PMC11653761. 🔗
-
Goh KJ, Altuvia Y, Argaman L, Raz Y, Bar A, Lithgow T, Margalit H, Gan YH. RIL-seq reveals extensive involvement of small RNAs in virulence and capsule regulation in hypervirulent Klebsiella pneumoniae. Nucleic Acids Res. 2024 Aug 27;52(15):9119-9138. doi: 10.1093/nar/gkae440. PMID: 38804271; PMCID: PMC11347178. 🔗
-
Plum MTW, Cheung HC, Iscar PR, Chen Y, Gan YH, Basler M. Burkholderia thailandensis uses a type VI secretion system to lyse protrusions without triggering host cell responses. Cell Host Microbe. 2024 May 8;32(5):676-692.e5. doi: 10.1016/j.chom.2024.03.013. Epub 2024 Apr 18. PMID: 38640929. 🔗
-
Tan YH, Arros P, Berríos-Pastén C, Wijaya I, Chu WHW, Chen Y, Cheam G, Mohamed Naim AN, Marcoleta AE, Ravikrishnan A, Nagarajan N, Lagos R, Gan YH. Hypervirulent Klebsiella pneumoniae employs genomic island encoded toxins against bacterial competitors in the gut. ISME J. 2024 Jan 8;18(1):wrae054. doi: 10.1093/ismejo/wrae054. Erratum in: ISME J. 2024 Jan 8;18(1):wrae143. doi: 10.1093/ismejo/wrae143. PMID: 38547398; PMCID: PMC11020217. 🔗
-
Gálvez-Silva M, Arros P, Berríos-Pastén C, Villamil A, Rodas PI, Araya I, Iglesias R, Araya P, Hormazábal JC, Bohle C, Chen Y, Gan YH, Chávez FP, Lagos R, Marcoleta AE. Carbapenem-resistant hypervirulent ST23 Klebsiella pneumoniae with a highly transmissible dual-carbapenemase plasmid in Chile. Biol Res. 2024 Mar 12;57(1):7. doi: 10.1186/s40659-024-00485-2. PMID: 38475927; PMCID: PMC10929235. 🔗
-
Chang KC, Nagarajan N, Gan YH. Short-chain fatty acids of various lengths differentially inhibit Klebsiella pneumoniae and Enterobacteriaceae species. mSphere. 2024 Feb 28;9(2):e0078123. doi: 10.1128/msphere.00781-23. Epub 2024 Feb 2. PMID: 38305176; PMCID: PMC10900885. 🔗
-
Li M, You K, Wang P, Hooi L, Chen Y, Siah A, Tan SB, Teo J, Ng OT, Marimuthu K, Venkatachalam I, Blasiak A, Chow EKH, Ho D and Gan YH. Discovery of Broad-Spectrum Repurposed Drug Combinations Against Carbapenem-Resistant Enterobacteriaceae (CRE) Through Artificial Intelligence (AI)-driven Platform. Advanced Therapeutics. 2024 Jan 12; 7(3):2300332. doi: 10.1002/adtp.202300332. 🔗
-
Rojas D, Marcoleta AE, Gálvez-Silva M, Varas MA, Díaz M, Hernández M, Vargas C, Nourdin-Galindo G, Koch E, Saldivia P, Vielma J, Gan YH, Chen Y, Guiliani N, Chávez FP. Inorganic Polyphosphate Affects Biofilm Assembly, Capsule Formation, and Virulence of Hypervirulent ST23 Klebsiella pneumoniae. ACS Infect Dis. 2024 Feb 9;10(2):606-623. doi: 10.1021/acsinfecdis.3c00509. Epub 2024 Jan 11. PMID: 38205780. 🔗
-
Chen Y, Yong M, Li M, Si Z, Koh CH, Lau P, Chang YW, Teo J, Chan-Park MB, Gan YH. A hydrophilic polyimidazolium antibiotic targeting the membranes of Gram-negative bacteria. J Antimicrob Chemother. 2023 Oct 3;78(10):2581-2590. doi: 10.1093/jac/dkad274. PMID: 37671807; PMCID: PMC10545527. 🔗
-
Chu WHW, Tan YH, Tan SY, Chen Y, Yong M, Lye DC, Kalimuddin S, Archuleta S, Gan YH. Acquisition of regulator on virulence plasmid of hypervirulent Klebsiella allows bacterial lifestyle switch in response to iron. mBio. 2023 Aug 31;14(4):e0129723. doi: 10.1128/mbio.01297-23. Epub 2023 Aug 2. PMID: 37530523; PMCID: PMC10470599. 🔗
-
Yong M, Kok ZY, Koh CH, Zhong W, Ng JT, Mu Y, Chan-Park MB, Gan YH. Membrane Potential-Dependent Uptake of Cationic Oligoimidazolium Mediates Bacterial DNA Damage and Death. Antimicrob Agents Chemother. 2023 May 17;67(5):e0035523. doi: 10.1128/aac.00355-23. Epub 2023 May 1. PMID: 37125913; PMCID: PMC10190574. 🔗
-
Kang JTL, Teo JJY, Bertrand D, Ng A, Ravikrishnan A, Yong M, Ng OT, Marimuthu K, Chen SL, Chng KR, Gan YH, Nagarajan N. Long-term ecological and evolutionary dynamics in the gut microbiomes of carbapenemase-producing Enterobacteriaceae colonized subjects. Nat Microbiol. 2022 Oct;7(10):1516-1524. doi: 10.1038/s41564-022-01221-w. Epub 2022 Sep 15. PMID: 36109646; PMCID: PMC9519440. 🔗
-
Goncalves RA, Ku JWK, Zhang H, Salim T, Oo G, Zinn AA, Boothroyd C, Tang RMY, Gan CL, Gan YH, and Lam YM. Copper-Nanoparticle-Coated Fabrics for Rapid and Sustained Antibacterial Activity Applications. CS Appl. Nano Mater. 2022 Aug 25; 5(9):12876-12886. doi: 10.1021/acsanm.2c02736. 🔗
-
Yong M, Chen Y, Oo G, Chang KC, Chu WHW, Teo J, Venkatachalam I, Thevasagayam NM, Sridatta PSR, Koh V, Marcoleta AE, Chen H, Nagarajan N, Kalisvar M, Ng OT, Gan YH. Dominant Carbapenemase-Encoding Plasmids in Clinical Enterobacterales Isolates and Hypervirulent Klebsiella pneumoniae, Singapore. Emerg Infect Dis. 2022 Aug;28(8):1578-1588. doi: 10.3201/eid2808.212542. PMID: 35876475; PMCID: PMC9328930. 🔗
-
Lim DRX, Chen Y, Ng LF, Gruber J, Gan YH. Glutathione catabolism by Enterobacteriaceae species to hydrogen sulphide adversely affects the viability of host systems in the presence of 5’fluorodeoxyuridine. Mol Microbiol. 2022 May;117(5):1089-1103. doi: 10.1111/mmi.14893. Epub 2022 Mar 22. PMID: 35279884; PMCID: PMC9313583. 🔗
-
Ku JWK, Marsh ST, Nai MH, Robinson KS, Teo DET, Zhong FL, Brown KA, Lim TC, Lim CT, Gan YH. Skin models for cutaneous melioidosis reveal Burkholderia infection dynamics at wound’s edge with inflammasome activation, keratinocyte extrusion and epidermal detachment. Emerg Microbes Infect. 2021 Dec;10(1):2326-2339. doi: 10.1080/22221751.2021.2011621. PMID: 34821529; PMCID: PMC8654412. 🔗
-
Ku JWK, Gan YH. New roles for glutathione: Modulators of bacterial virulence and pathogenesis. Redox Biol. 2021 Aug;44:102012. doi: 10.1016/j.redox.2021.102012. Epub 2021 May 29. PMID: 34090244; PMCID: PMC8182430. 🔗
-
Molton JS, Lee IR, Bertrand D, Ding Y, Kalimuddin S, Lye DC, Nagarajan N, Gan YH, Archuleta S. Stool metagenome analysis of patients with Klebsiella pneumoniae liver abscess and their domestic partners. Int J Infect Dis. 2021 Jun;107:1-4. doi: 10.1016/j.ijid.2021.04.012. Epub 2021 Apr 16. PMID: 33862216. 🔗
-
Zhong W, Shi Z, Mahadevegowda SH, Liu B, Zhang K, Koh CH, Ruan L, Chen Y, Zeden MS, Pee CJE, Marimuthu K, De PP, Ng OT, Zhu Y, Chi YR, Hammond PT, Yang L, Gan YH, Pethe K, Greenberg EP, Gründling A, Chan-Park MB. Designer broad-spectrum polyimidazolium antibiotics. Proc Natl Acad Sci U S A. 2020 Dec 8;117(49):31376-31385. doi: 10.1073/pnas.2011024117. Epub 2020 Nov 23. PMID: 33229526; PMCID: PMC7739875. 🔗
-
Hoong BYD, Gan YH, Liu H, Chen ES. cGAS-STING pathway in oncogenesis and cancer therapeutics. Oncotarget. 2020 Jul 28;11(30):2930-2955. doi: 10.18632/oncotarget.27673. PMID: 32774773; PMCID: PMC7392626. 🔗
-
Ku JWK, Chen Y, Lim BJW, Gasser S, Crasta KC, Gan YH. Bacterial-induced cell fusion is a danger signal triggering cGAS-STING pathway via micronuclei formation. Proc Natl Acad Sci U S A. 2020 Jul 7;117(27):15923-15934. doi: 10.1073/pnas.2006908117. Epub 2020 Jun 22. PMID: 32571920; PMCID: PMC7355030. 🔗
-
Chng KR, Ghosh TS, Tan YH, Nandi T, Lee IR, Ng AHQ, Li C, Ravikrishnan A, Lim KM, Lye D, Barkham T, Raman K, Chen SL, Chai L, Young B, Gan YH, Nagarajan N. Metagenome-wide association analysis identifies microbial determinants of post-antibiotic ecological recovery in the gut. Nat Ecol Evol. 2020 Sep;4(9):1256-1267. doi: 10.1038/s41559-020-1236-0. Epub 2020 Jul 6. PMID: 32632261. 🔗
-
Qi G, Hu F, Kenry, Chong KC, Wu M, Gan YH, and Liu B. Bacterium‐Templated Polymer for Self‐Selective Ablation of Multidrug‐Resistant Bacteria. Advanced Functional Materials. 2020 Jun; 30 (31): 2001338. doi: 10.1002/adfm.202001338. 🔗
-
Chen Y, Marimuthu K, Teo J, Venkatachalam I, Cherng BPZ, De Wang L, Prakki SRS, Xu W, Tan YH, Nguyen LC, Koh TH, Ng OT, Gan YH. Acquisition of Plasmid with Carbapenem-Resistance Gene blaKPC2 in Hypervirulent Klebsiella pneumoniae, Singapore. Emerg Infect Dis. 2020 Mar;26(3):549-559. doi: 10.3201/eid2603.191230. PMID: 32091354; PMCID: PMC7045839. 🔗
-
Si Z, Lim HW, Tay MYF, Du Y, Ruan L, Qiu H, Zamudio-Vazquez R, Reghu S, Chen Y, Tiong WS, Marimuthu K, De PP, Ng OT, Zhu Y, Gan YH, Chi YR, Duan H, Bazan GC, Greenberg EP, Chan-Park MB, Pethe K. A Glycosylated Cationic Block Poly(β-peptide) Reverses Intrinsic Antibiotic Resistance in All ESKAPE Gram-Negative Bacteria. Angew Chem Int Ed Engl. 2020 Apr 20;59(17):6819-6826. doi: 10.1002/anie.201914304. Epub 2020 Feb 19. PMID: 32011781. 🔗
-
Tan YH, Chen Y, Chu WHW, Sham LT, Gan YH. Cell envelope defects of different capsule-null mutants in K1 hypervirulent Klebsiella pneumoniae can affect bacterial pathogenesis. Mol Microbiol. 2020 May;113(5):889-905. doi: 10.1111/mmi.14447. Epub 2020 Jan 20. PMID: 31912541; PMCID: PMC7317392. 🔗
-
Hoh CH, Tan YH, Gan YH. Protective Role of Kupffer Cells and Macrophages in Klebsiella pneumoniae-Induced Liver Abscess Disease. Infect Immun. 2019 Aug 21;87(9):e00369-19. doi: 10.1128/IAI.00369-19. PMID: 31285251; PMCID: PMC6704610. 🔗
-
Ku JWK, Gan YH. Modulation of bacterial virulence and fitness by host glutathione. Curr Opin Microbiol. 2019 Feb;47:8-13. doi: 10.1016/j.mib.2018.10.004. Epub 2018 Nov 2. PMID: 30396015. 🔗
-
Lam MMC, Wyres KL, Duchêne S, Wick RR, Judd LM, Gan YH, Hoh CH, Archuleta S, Molton JS, Kalimuddin S, Koh TH, Passet V, Brisse S, Holt KE. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun. 2018 Jul 13;9(1):2703. doi: 10.1038/s41467-018-05114-7. PMID: 30006589; PMCID: PMC6045662. 🔗
-
Gamage AM, Liao C, Cheah IK, Chen Y, Lim DRX, Ku JWK, Chee RSL, Gengenbacher M, Seebeck FP, Halliwell B, Gan YH. The proteobacterial species Burkholderia pseudomallei produces ergothioneine, which enhances virulence in mammalian infection. FASEB J. 2018 Jun 11:fj201800716. doi: 10.1096/fj.201800716. Epub ahead of print. PMID: 29890088. 🔗
-
Lee IR, Sng E, Lee KO, Molton JS, Chan M, Kalimuddin S, Izharuddin E, Lye DC, Archuleta S, Gan YH. Comparison of Diabetic and Non-diabetic Human Leukocytic Responses to Different Capsule Types of Klebsiella pneumoniae Responsible for Causing Pyogenic Liver Abscess. Front Cell Infect Microbiol. 2017 Sep 7;7:401. doi: 10.3389/fcimb.2017.00401. PMID: 28936426; PMCID: PMC5594087. 🔗
-
Gamage AM, Lee KO, Gan YH. Anti-Cancer Drug HMBA Acts as an Adjuvant during Intracellular Bacterial Infections by Inducing Type I IFN through STING. J Immunol. 2017 Oct 1;199(7):2491-2502. doi: 10.4049/jimmunol.1602162. Epub 2017 Aug 21. PMID: 28827286. 🔗
-
Tan YH, Gamage AM, Gan YH. Complement-activated vitronectin enhances the invasion of nonphagocytic cells by bacterial pathogens Burkholderia and Klebsiella. Cell Microbiol. 2017 Aug;19(8). doi: 10.1111/cmi.12732. Epub 2017 Mar 21. PMID: 28186697. 🔗
-
Lee IR, Molton JS, Wyres KL, Gorrie C, Wong J, Hoh CH, Teo J, Kalimuddin S, Lye DC, Archuleta S, Holt KE, Gan YH. Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci Rep. 2016 Jul 13;6:29316. doi: 10.1038/srep29316. PMID: 27406977; PMCID: PMC4942785. 🔗
-
Wong J, Chen Y, Gan YH. Host Cytosolic Glutathione Sensing by a Membrane Histidine Kinase Activates the Type VI Secretion System in an Intracellular Bacterium. Cell Host Microbe. 2015 Jul 8;18(1):38-48. doi: 10.1016/j.chom.2015.06.002. Epub 2015 Jun 18. PMID: 26094804. 🔗
-
Lim YT, Jobichen C, Wong J, Limmathurotsakul D, Li S, Chen Y, Raida M, Srinivasan N, MacAry PA, Sivaraman J, Gan YH. Extended loop region of Hcp1 is critical for the assembly and function of type VI secretion system in Burkholderia pseudomallei. Sci Rep. 2015 Feb 4;5:8235. doi: 10.1038/srep08235. PMID: 25648885; PMCID: PMC4650826. 🔗
-
Teh BE, French CT, Chen Y, Chen IG, Wu TH, Sagullo E, Chiou PY, Teitell MA, Miller JF, Gan YH. Type three secretion system-mediated escape of Burkholderia pseudomallei into the host cytosol is critical for the activation of NFκB. BMC Microbiol. 2014 May 6;14:115. doi: 10.1186/1471-2180-14-115. PMID: 24884837; PMCID: PMC4026835. 🔗
-
Gamage AM, Lee KO, Gan YH. Effect of oral N-acetyl cysteine supplementation in type 2 diabetic patients on intracellular glutathione content and innate immune responses to Burkholderia pseudomallei. Microbes Infect. 2014 Aug;16(8):661-71. doi: 10.1016/j.micinf.2014.07.007. Epub 2014 Aug 1. PMID: 25088507. 🔗
-
Chen Y, Schröder I, French CT, Jaroszewicz A, Yee XJ, Teh BE, Toesca IJ, Miller JF, Gan YH. Characterization and analysis of the Burkholderia pseudomallei BsaN virulence regulon. BMC Microbiol. 2014 Aug 1;14:206. doi: 10.1186/s12866-014-0206-6. PMID: 25085508; PMCID: PMC4236580. 🔗
-
Ooi WF, Ong C, Nandi T, Kreisberg JF, Chua HH, Sun G, Chen Y, Mueller C, Conejero L, Eshaghi M, Ang RM, Liu J, Sobral BW, Korbsrisate S, Gan YH, Titball RW, Bancroft GJ, Valade E, Tan P. The condition-dependent transcriptional landscape of Burkholderia pseudomallei. PLoS Genet. 2013;9(9):e1003795. doi: 10.1371/journal.pgen.1003795. Epub 2013 Sep 12. PMID: 24068961; PMCID: PMC3772027. 🔗
-
Lee SH, Wong RR, Chin CY, Lim TY, Eng SA, Kong C, Ijap NA, Lau MS, Lim MP, Gan YH, He FL, Tan MW, Nathan S. Burkholderia pseudomallei suppresses Caenorhabditis elegans immunity by specific degradation of a GATA transcription factor. Proc Natl Acad Sci U S A. 2013 Sep 10;110(37):15067-72. doi: 10.1073/pnas.1311725110. Epub 2013 Aug 26. PMID: 23980181; PMCID: PMC3773767. 🔗
-
Gan YH. Host susceptibility factors to bacterial infections in type 2 diabetes. PLoS Pathog. 2013;9(12):e1003794. doi: 10.1371/journal.ppat.1003794. Epub 2013 Dec 26. PMID: 24385898; PMCID: PMC3873456. 🔗
-
Tan KS, Lee KO, Low KC, Gamage AM, Liu Y, Tan GY, Koh HQ, Alonso S, Gan YH. Glutathione deficiency in type 2 diabetes impairs cytokine responses and control of intracellular bacteria. J Clin Invest. 2012 Jun;122(6):2289-300. doi: 10.1172/JCI57817. Epub 2012 May 1. PMID: 22546856; PMCID: PMC3366396. 🔗
-
Chen Y, Wong J, Sun GW, Liu Y, Tan GY, Gan YH. Regulation of type VI secretion system during Burkholderia pseudomallei infection. Infect Immun. 2011 Aug;79(8):3064-73. doi: 10.1128/IAI.05148-11. Epub 2011 Jun 13. PMID: 21670170; PMCID: PMC3147588. 🔗
-
Sun GW, Gan YH. Unraveling type III secretion systems in the highly versatile Burkholderia pseudomallei. Trends Microbiol. 2010 Dec;18(12):561-8. doi: 10.1016/j.tim.2010.09.002. Epub 2010 Oct 15. PMID: 20951592. 🔗
-
Sun GW, Chen Y, Liu Y, Tan GY, Ong C, Tan P, Gan YH. Identification of a regulatory cascade controlling Type III Secretion System 3 gene expression in Burkholderia pseudomallei. Mol Microbiol. 2010 May;76(3):677-89. doi: 10.1111/j.1365-2958.2010.07124.x. Epub 2010 Mar 24. PMID: 20345664. 🔗
-
Tan KS, Chen Y, Lim YC, Tan GY, Liu Y, Lim YT, Macary P, Gan YH. Suppression of host innate immune response by Burkholderia pseudomallei through the virulence factor TssM. J Immunol. 2010 May 1;184(9):5160-71. doi: 10.4049/jimmunol.0902663. Epub 2010 Mar 24. PMID: 20335533. 🔗
-
Lee YH, Chen Y, Ouyang X, Gan YH. Identification of tomato plant as a novel host model for Burkholderia pseudomallei. BMC Microbiol. 2010 Jan 29;10:28. doi: 10.1186/1471-2180-10-28. PMID: 20109238; PMCID: PMC2823722. 🔗
-
Breitbach K, Sun GW, Köhler J, Eske K, Wongprompitak P, Tan G, Liu Y, Gan YH, Steinmetz I. Caspase-1 mediates resistance in murine melioidosis. Infect Immun. 2009 Apr;77(4):1589-95. doi: 10.1128/IAI.01257-08. Epub 2009 Jan 29. PMID: 19179418; PMCID: PMC2663179. 🔗
-
Ye Z, Lee CM, Sun GW, Gan YH. Burkholderia pseudomallei infection of T cells leads to T-cell costimulation partially provided by flagellin. Infect Immun. 2008 Jun;76(6):2541-50. doi: 10.1128/IAI.01310-07. Epub 2008 Mar 17. PMID: 18347031; PMCID: PMC2423057. 🔗
-
Hii CS, Sun GW, Goh JW, Lu J, Stevens MP, Gan YH. Interleukin-8 induction by Burkholderia pseudomallei can occur without Toll-like receptor signaling but requires a functional type III secretion system. J Infect Dis. 2008 Jun 1;197(11):1537-47. doi: 10.1086/587905. PMID: 18419546. 🔗
-
Ye Z, Gan YH. Flagellin contamination of recombinant heat shock protein 70 is responsible for its activity on T cells. J Biol Chem. 2007 Feb 16;282(7):4479-4484. doi: 10.1074/jbc.M606802200. Epub 2006 Dec 18. PMID: 17178717. 🔗
-
Koo GC, Gan YH. The innate interferon gamma response of BALB/c and C57BL/6 mice to in vitro Burkholderia pseudomallei infection. BMC Immunol. 2006 Aug 18;7:19. doi: 10.1186/1471-2172-7-19. PMID: 16919160; PMCID: PMC1559720. 🔗
-
Gan YH. Interaction between Burkholderia pseudomallei and the host immune response: sleeping with the enemy? J Infect Dis. 2005 Nov 15;192(10):1845-50. doi: 10.1086/497382. Epub 2005 Oct 7. PMID: 16235187. 🔗
-
Sun GW, Lu J, Pervaiz S, Cao WP, Gan YH. Caspase-1 dependent macrophage death induced by Burkholderia pseudomallei. Cell Microbiol. 2005 Oct;7(10):1447-58. doi: 10.1111/j.1462-5822.2005.00569.x. PMID: 16153244. 🔗
-
Chen K, Sun GW, Chua KL, Gan YH. Modified virulence of antibiotic-induced Burkholderia pseudomallei filaments. Antimicrob Agents Chemother. 2005 Mar;49(3):1002-9. doi: 10.1128/AAC.49.3.1002-1009.2005. PMID: 15728895; PMCID: PMC549247. 🔗
-
Chua KL, Chan YY, Gan YH. Flagella are virulence determinants of Burkholderia pseudomallei. Infect Immun. 2003 Apr;71(4):1622-9. doi: 10.1128/IAI.71.4.1622-1629.2003. PMID: 12654773; PMCID: PMC152022. 🔗
-
Gan YH, Chua KL, Chua HH, Liu B, Hii CS, Chong HL, Tan P. Characterization of Burkholderia pseudomallei infection and identification of novel virulence factors using a Caenorhabditis elegans host system. Mol Microbiol. 2002 Jun;44(5):1185-97. doi: 10.1046/j.1365-2958.2002.02957.x. PMID: 12068805. 🔗
-
Liu B, Koo GC, Yap EH, Chua KL, Gan YH. Model of differential susceptibility to mucosal Burkholderia pseudomallei infection. Infect Immun. 2002 Feb;70(2):504-11. doi: 10.1128/IAI.70.2.504-511.2002. PMID: 11796576; PMCID: PMC127661. 🔗

